Abstract
Introduction: Beta three adrenergic receptor (ADRB3) is an adrenergic receptor that induces activation of adenylate cyclase located mainly in adipose tissue and is involved in the thermogenesis of brown fat tissue and in the regulation of lipolysis. Agonists of ADRB3 are found to induce the thermogenesis process of human brown fat tissue and thus believed to be excellent anti-obesity targets. The most studied single nucleotide polymorphism (SNP) of ADRB3 is rs4994. Inconsistent findings have been found in earlier studies about the association of rs4994 polymorphisms with obesity among different populations. The association of ADRB3/rs4994 polymorphism with obesity among the Saudi population is unknown. This study aimed to investigate the association of ADRB3/rs4994 polymorphism with obesity, blood lipids and blood pressure in the Saudi population.
Method: This study was a case control study involving 88 obese healthy volunteers and 84 non-obese (controls) volunteers recruited from the King Khaled University Hospital (KKUH), Riyadh City, Saudi Arabia. Using KASPTM (Competitive Allele-Specific PCR) the rs4994 genotype for each participant was determined. The frequency, distribution, and association of each genotype with body mass index (BMI) and lipid profile were calculated.
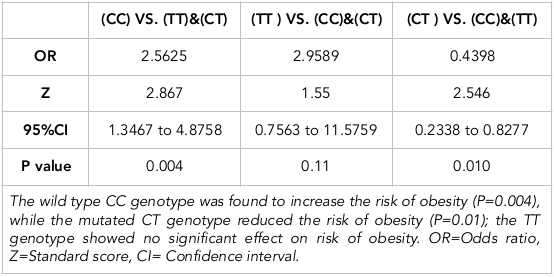
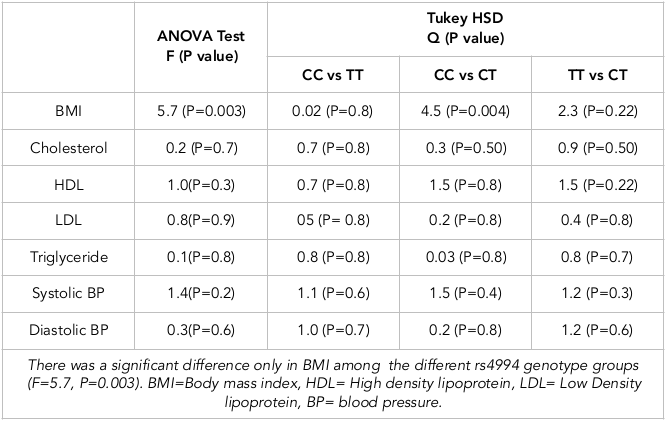
Results: The distribution of CC, TT and CT genotypes in the study population was 0.37, 0.06 and 0.56, respectively. The heterozygote CT genotype was associated with a reduced risk of obesity (odds ratio (OR)=0.4398, 95%CI=0.2338 to 0.8277, P-value=0.010). It was more frequent in the non-obese participants compared to the obese participants (67.9% vs. 44.3%, respectively). Moreover, participants with the CT genotype had a significantly lower BMI (P=0.004). In contrast, the CC genotype was associated with an increased risk of obesity (OR=2.5, 95%CI=1.3467 to 4.8758, P-value=0.004). The frequency of the CC genotype was higher in obese participants compared to the non-obese ones (46.6% vs. 28.6%, respectively). Participants with the CC genotype demonstrated a significantly higher BMI than participants with the CT or TT genotypes (Q= 4.5, P=0.004). The TT genotype had no significant effects on the participants’ BMI (OR=2.9, 95%CI=0.7563 to 11.5759, P value=0.11), and it was higher in obese compared to non-obese participants (9.1% vs. 3.6%, respectively). No significant effect of ADRB3/rs4994 polymorphism on blood lipid profile or blood pressure was observed.
Conclusion: The findings of this study suggested that the heterozygote CT genotype of the ADRB3/rs4994 polymorphism is associated with a reduced risk of obesity among the Saudi population. In the future, larger scale studies are required to further confirm these observations.
Introduction
The beta three adrenergic receptor (ADRB3) is one of the beta adrenergic receptors that can induce the activation of adenylate cyclase through the action of G proteins. It is located mainly in adipose tissues; however, it is also involved in the regulation of lipolysis and thermogenesis. ADRB3 is believed to be involved in glucose and lipid metabolism, and in the thermogenesis process, thus directly or indirectly influencing body weight regulation. Agonists of ADRB3 are found to induce human brown fat tissue thermogenesis and thus believed to be excellent anti-obesity therapy targets Cypess et al., 2015.
The most studied single nucleotide polymorphism (SNP) of ADRB3 is rs4994 (Trp64Arg), a missense mutation that replaces tryptophan with arginine Mirrakhimov et al., 2011. It was hypothesized that this mutation reduces the affinity of ADRB3 to norepinephrine in adipocytes. An interesting study found that the rs4994 mutation reduces lipolytic activity of ADRB3 in human adipose cells Umekawa et al., 1999. The association of ADRB3/rs4994 polymorphism with obesity is still inconsistent from earlier studies. The polymorphism was found to be positively associated with obesity in some studies Fujisawa et al., 1998Kurokawa et al., 2001Szendrei et al., 2016. However, the polymorphism was not associated with obesity in other studies Liu et al., 2015Shiwaku et al., 2003Zawodniak-Szalapska et al., 2008.
Indeed, the polymorphism was associated with protection against obesity in the Brazilian population (odds ratio (OR)=0; P=0.01) Brondani et al., 2014, in the African American population McKean-Cowdin et al., 2007, and in the Tatar population Kochetova et al., 2015. As well, it was associated with increasing athletic endurance performance Santiago et al., 2011. These variations in findings could be explained by the variations in the ethnic backgrounds of the studied populations. Interestingly, a meta-analysis study suggested that the mutation increases the risk of obesity in East Asian populations but not in European ones Kurokawa et al., 2008.
Moreover, obesity is a significant health problem among the Saudi population, affecting 29% of the population Memish et al., 2014. The genetic factors seem to play an important role in obesity among Saudi populations since we observed a familial clustering Al Dahi et al., 2014Shaikh et al., 2016. Interestingly, an earlier study by Abu Amero et al. (2005) found that the rs4994 mutation was not associated with risk of coronary artery diseases among Saudi patients Abu-Amero et al., 2005.
Few studies have been conducted to assess the genetic factors contributing to obesity among the Saudi population and, thus, further studies are required. The ADRB3/rs4994 is a potential risk locus for obesity. However, this gene mutation has been found to be important in inducing non-shivering thermogenesis in brown fat tissues. Thus, alteration in this gene could cut energy expenditure and significantly promote obesity. Understanding the genetic loci associated with risk of obesity will lead to a better understanding of the underlying pathophysiological mechanisms. This should lead to better management and effective prevention of obesity among the Saudi population.
The current study was aimed at evaluating the association of the ADRB3/rs4994 polymorphism with obesity (and blood lipid profile) among Saudis.
Materials-Methods
Recruitment of participants
We conducted a case control study involving 88 obese individuals and 84 non-obese individuals (controls) recruited from healthy volunteers visiting the King Khaled University Hospital (KKUH), Riyadh City, Saudi Arabia. The inclusion criteria included: Saudi nationality, adult age (above 18 years old), and body mass index (BMI) of greater than 30 (for obese group) or less than 30 (for non-obese group). The participants with current acute or chronic illnesses were excluded. Ethical approval of the study protocol was provided by the Institutional Review Board (IRB) of the College of Medicine, King Saud University (Reference number of 16/0283/IRB). After explaining the procedure with the participants, as well as the purpose of the study and potential risks associated with participation, an informed consent (approved by the IRB) was obtained from each participant. The anthropometric measurements (weight and height) and blood pressure were measured for each participant using electronic medical scales available at the KKUH.
Sample collection
Venous blood was drawn from each participant by an expert phlebotomist. About 2 mL of the extracted blood was placed in an EDTA tube, and 3 mL was placed in a gel separator tube. The tubes were then transferred on ice immediately to the College of Medicine Research Center (CMRC). However, the EDTA tubes were stored at 4°C until use for DNA extraction. The gel separator tubes were centrifuged at 4°C for 15 min at 1500 g within 30 min to obtain serum. The serum was allocated and stored at -80°C until use in biochemical analysis.
DNA extraction
The acid guanidinium thiocyanate-phenol-chloroform extraction method was used to obtain DNA from venous blood stored in EDTA tubes. In brief, 0.5 mL of blood was added to 0.5 mL of TRIGentTM (Biomatik, USA, Cat. No. K5161). After 10 minutes of incubation at room temperature with intermittent vortexing, 100 μL of chloroform was added. Then, after a 5-min incubation at room temperature the tube was centrifuged for 10-15 min at 12,000 g, and the aqueous upper layer was removed. Then, 0.5 mL of 100% ethanol was added to the lower organic phase to precipitate the DNA. To obtain the DNA pellet, the tubes were centrifuged at 4°C for 10 min at highest speed. The DNA pellets were washed twice using 70% ethanol and air dried for 10 to 15 min. PCR-grade water was used to dissolve the DNA pellets. The DNA quality and quantity were measured using the Nano Drop ND-1000 UV-VIS spectrophotometer (version 3.2.1). The DNA was then stored at -80° until use for genotyping.
Single Nucleotide Polymorphism (SNP) genotyping
The selected SNP was genotyped using KASPTM (Competitive Allele-Specific PCR) method developed by Kbiosciences, UK. Details of the method and concepts of the KASPTM are provided on the manufacturer’s website (Kbiosciences, UK). In brief, the DNA samples were diluted to be less than 50 ng/μL. Then the diluted DNA samples were arrayed into 96-well plates. The reaction mix for each well was added, containingEDTA of PCR grade water and 5 μL of KASPTM genotyping mix (the SNP assay was added prior using a ratio of 0.14: 5 for assay to genotype mix). The real time PCR reaction was run in a PCR machine (Applied Biosystems™ ViiA™ 7 System, Applied Biosystems, USA) available at the Stem Cell Research Center, College of Medicine, King Saud University. The thermal cycle reaction was as follows:
Activation, 15 minutes at 94° (one cycle)
First PCR cycles, denaturing at 94° for 20 seconds followed by annealing/elongation at 60° with cool-down of 0.6° for 60 seconds (10 cycles).
Second PCR cycles, denaturing at 94° for 20 seconds followed by annealing/ elongation at 55° for 60 seconds (up to 33 cycles).
Finally, the ensued signal was read at 35° for 1 minute and the genotypes were discerned automatically by the real time PCR machine.
Blood lipid profile
The blood lipid profile was measured in the participants’ serum using the Dimension VistaTM 1500 System available in the KKUH laboratory department.
Statistical analysis
The data were presented as mean and standard deviation. The frequency of each genotype was compared between the obese vs. non-obese groups. The association of each genotype with obesity, blood lipid profile, or blood pressure was calculated using odds ratio (OR). The difference between the groups was assessed using analysis of variance (Anova) test followed by post-hoc Tukey honest significant difference (HSD) test. Any difference was considered significant if P<0.05. The SPSS program version 20 (Statistical Package for the Social Sciences, USA) was used for analyses.
Results
Characteristics of study participants
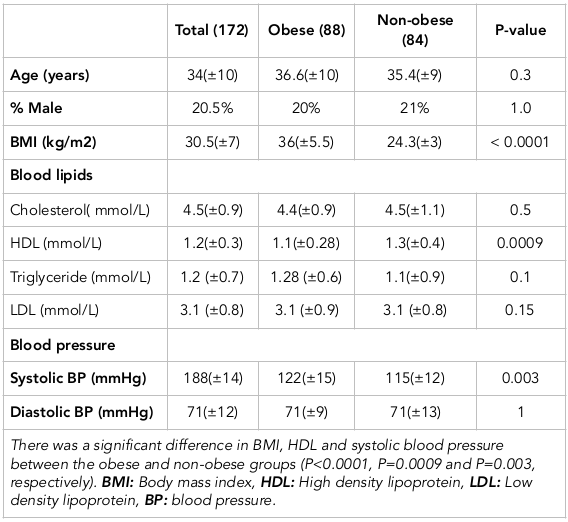
The characteristics of the study participants are presented in Table 1 . A total of 172 participants were involved in the study; 88 were obese (BMI >30) and 84 were non-obese (BMI <30). The mean BMI was 36 kg/m2 (±5.5) in the obese group and 24 kg/m2 (±3) in the non-obese group (p<0.0001). The two groups were comparable in age and sex (p=0.3, p=1.0, respectively). Interestingly, we found that the non-obese participants had a higher high density lipoprotein (HDL) level compared to the obese group (1.3 mmol/L vs. 1.1 mmol/L, respectively; p=0.0009). However, we found no significant difference in blood cholesterol, low density lipoprotein (LDL) or blood triglyceride levels among the two groups (p =0.5 and p=0.1, respectively). We found that obese participants had a higher systolic blood pressure compared to the non-obese group (122 mmHg vs. 115 mmHg, respectively; p=0.003) but there was no significant difference in diastolic blood pressure (p=1.0).
Association of ADRB3/rs4994 with BMI
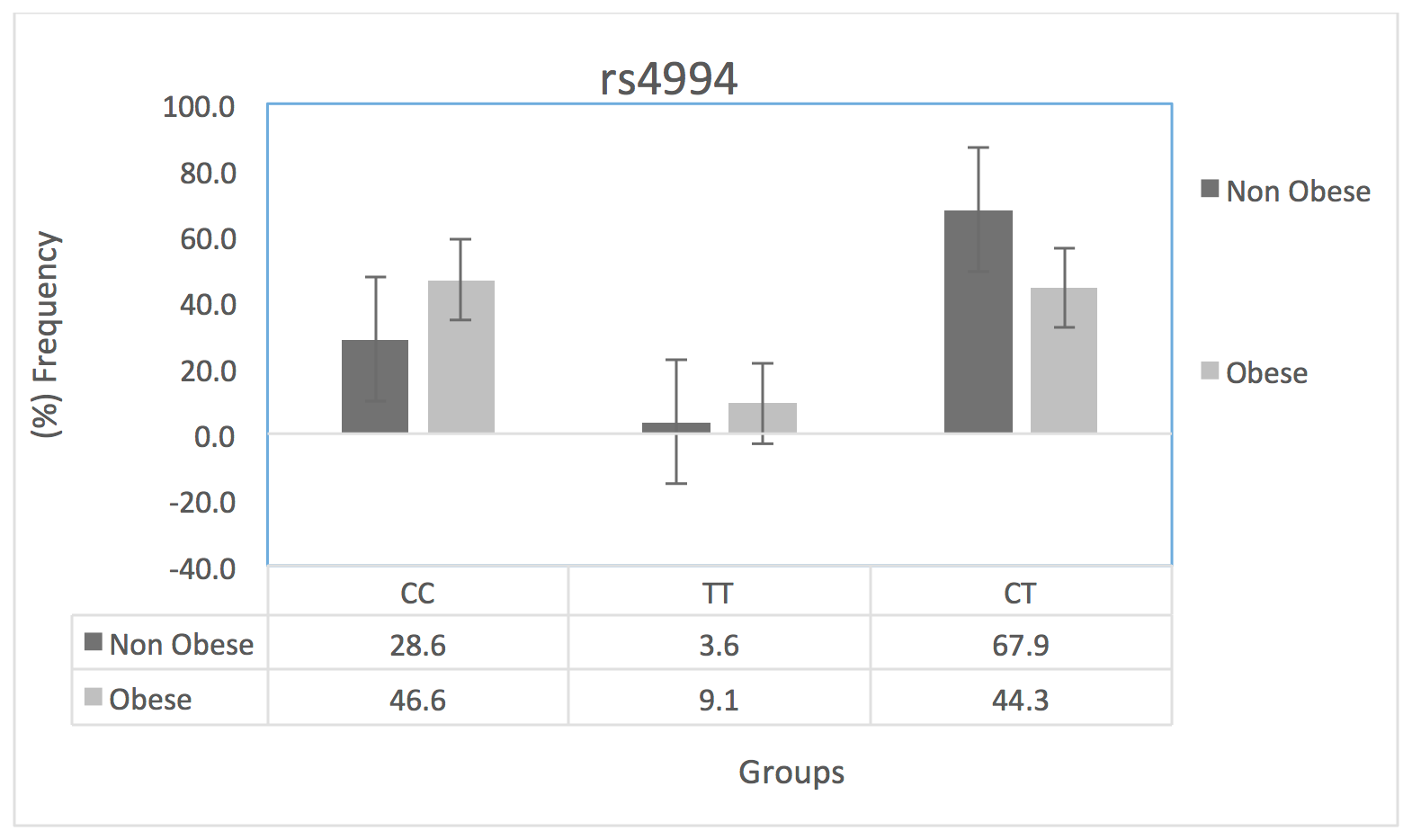
We aimed to assess the association of ADRB3/rs4994 with obesity reflected by BMI and blood lipid profile among Saudis. The ADRB3/rs4994 polymorphism was genotyped for each participant using the KASPTM method (Kbiosciences, UK). Then, the participants in each group (obese and non-obese groups) were further sub-grouped according to their ADRB3/rs4994 genotype. The distribution of CC, TT and CT genotypes in the study population was 0.37, 0.06 and 0.56, respectively. This distribution did not deviate from the Hardy-Weinberg equilibrium significantly (p=0.9). The chi-square random variable (Χ2) of Fisher’s exact test was 10.0 (p=0.006), indicating a dependence of the participants’ BMI on the ADRB3/rs4994 genotype.
We assessed the distribution of each ADRB3/rs4994 genotype among obese vs. non-obese participants. It was clear from our results that the distribution of different ADRB3/rs4994 genotypes was not comparable between the obese and non-obese groups. As shown in Figure 1 , the frequency of the CT genotype was the highest among all ADRB3/rs4994 genotypes. However, the frequency of this genotype was found to be highest in the non-obese participants compared to obese participants (67.9 % vs. 44.3% , respectively), suggesting the protective effect of this genotype. The TT genotype was found to be the least frequent; it was higher in obese than non-obese participants (9.1% vs. 3.6%, respectively). Meanwhile, the CC genotype was higher in obese participants compared to non-obese (46.6% vs. 28.6%, respectively).
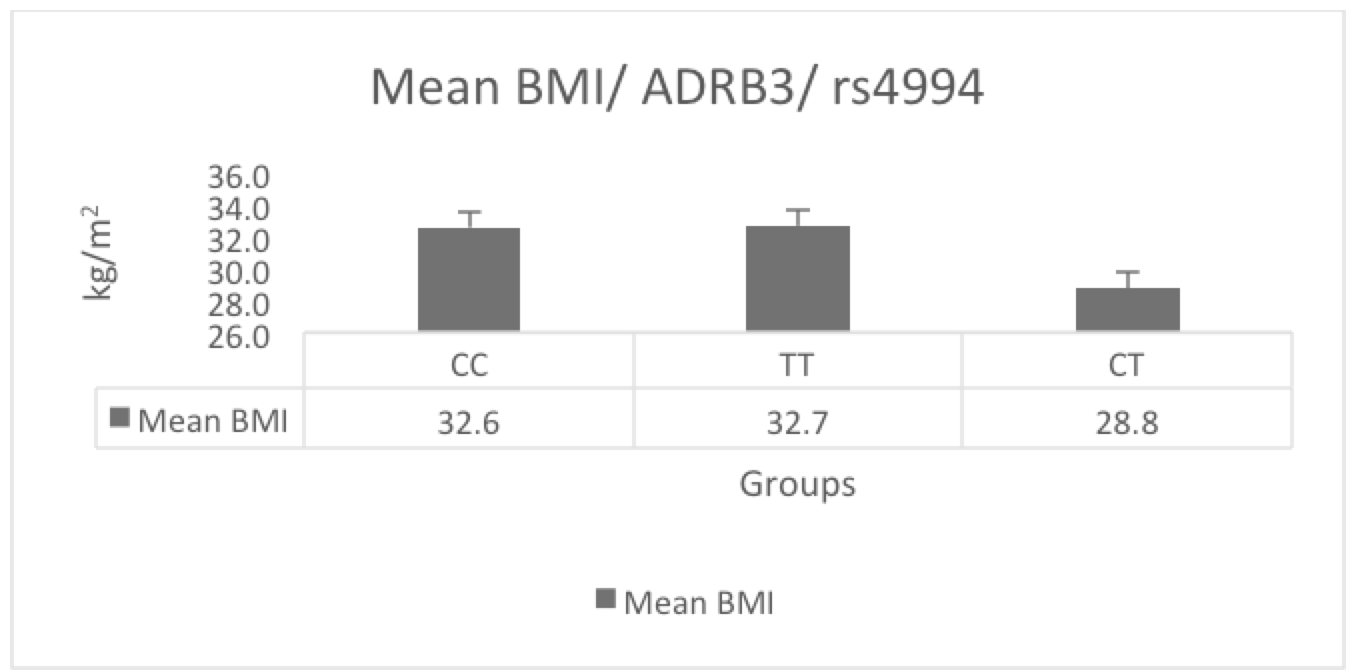
Furthermore, as displayed in Figure 2 , we assessed the mean BMI in the three genotype groups. Participants with CT genotype had a lower BMI mean (28.8 kg/m2), compared to participants with CC and TT genotypes (32.6 kg/m2 and 32.7 kg/m2, respectively), suggesting the protective effect of this genotype. We calculated the risk of obesity associated with each genotype using OR. As shown in Table 2 , we found that the CT genotype had a significant effect on reducing BMI; it was associated with a 57% reduction of obesity (OR=0.4398, 95%CI=0.2338 to 0.8277, P-value=0.010). In contrast, the CC genotype was associated with an increased risk of obesity (OR=2.5, 95%CI=1.3467 to 4.8758, P-value=0.004). Moreover, the TT genotype showed no significant effects on the participants’ BMI probably due to its low frequency (OR=2.9, 95%CI=0.7563 to 11.5759, P-value=0.11); the statistical power of this association analysis was estimated to be 0.83 Bacchetti & Leung, 2002.
Table 3 demonstrates the results of the ANOVA test analysis followed by post-hoc Tukey HSD test, which were used to assess the differences in BMI, blood lipid profile and blood pressure among the ADRB3/rs4994 genotypes groups. The ANOVA test showed a significant difference only in BMI among the groups (F=5.7, P=0.003). Furthermore, using post-hoc Tukey HSD testing, as demonstrated in Table 3, participants with the CC genotype had a significantly higher BMI than participants with the CT genotype (Q=4.5, P=0.004).
Association of ADRB3/rs4994 with blood lipids and blood pressure
We assessed the association of the ADRB3/rs4994 genotypes with blood lipid profile (cholesterol, HDL, and triglycerides). We found that participants with the TT genotype had a lower blood cholesterol mean (3.9 mmol/L), while other genotypes (CC and CT) had higher mean (4.45 mmol/L and 4.5 mmol/L, respectively). As shown in Table 3 , using ANOVA test and post-hoc Tukey HSD analysis, we found no significant differences in blood cholesterol levels among the groups (F=0.2, P= 0.7). Also, there was no significant difference in HDL (F=1.0, P= 0.3), LDL (F=0.8, P= 0.9), or triglycerides (F=0.1, P= 0.8) among the groups.
Interestingly, participants with CT genotype had a higher HDL mean (1.27 mmol/L) compared to AA and AG genotypes (1.14 mmol/L and 1.11 mmol/L, respectively). The student’s t test showed that participants with the CT genotype had a marginally significant higher blood HDL than participants of the other groups (t= 1.3, p= 0.09). Also, participants with the TT genotype had a relatively higher mean of HDL (1.28 mmol/L) compared to the CC and CT genotypes (1.20 mmol/L and 1.21 mmol/L, respectively).
Moreover, using ANOVA test and post-hoc Tukey HSD analysis we found no significant difference in systolic or diastolic blood pressure levels among the groups (F=1.4 P=0.2 and F= 0.3, P =0.6, respectively). The mean systolic blood pressure in the CC group was 117 mmHg, compared to 110 mmHg and 114 mmHg in CC and CT groups, respectively. Student’s t test showed that the CT group had a reduced systolic blood pressure which was marginally significant (t=-1.6, p= 0.06). The mean diastolic blood pressure was lower in participants of the TT group (66 mmHg), compared to participants in the CC and CT groups (69 mmHg).
Discussion
ADRB3 is involved in the regulation of thermogenesis and lipolysis in fat tissues. It was found to be involved in glucose and lipid metabolism, and in the thermogenesis process, thus directly or indirectly influencing body weight regulation. The missense mutation rs4994 is the most studied ADRB3 polymorphism. Earlier studies on the role of the rs4994 polymorphism with obesity have been inconsistent and lacking in the Saudi population. The distribution of CC, TT and CT genotypes in this study population was 0.37, 0.06 and 0.56, respectively. The frequency of the CT genotype was higher than observed in other populations de Luis et al., 2007Morcillo et al., 2008Takeuchi et al., 2012. Moreover, it was higher than observed previously in Saudis Abu-Amero et al., 2005.
Our study demonstrated that the wildtype CC genotype of rs4994 was associated with an increased risk of obesity (OR=2.5, 95%CI=1.3467 to 4.8758, P-value= 0.004). The frequency of the CC genotype was higher in obese participants compared to non-obese ones (46.6% vs. 28.6%, respectively). Additionally, participants with the CC genotype showed a significantly higher BMI mean than participants with the CT genotype (Q=4.5, P= 0.004).
On the other hand, the CT genotype was associated with a reduced risk of obesity (OR=0.4398, 95%CI=0.2338 to 0.8277, P-value=0.010). The frequency of this genotype was found to be high in the non-obese participants compared to the obese participants (67.9% vs. 44.3%, respectively). Also, participants with the CT genotype had a significantly lower BMI mean (p=0.004). Meanwhile the mutated TT genotype showed no significant effects on participants’ BMI (OR=2.9, 95%CI=0.7563 to 11.5759, P-value=0.11), probably due to its low frequency; thus, it was non-significant. It was more frequent in obese individuals compared to non-obese ones (9.1% vs. 3.6%, respectively).
Our findings are consistent with earlier studies of the Brazilian population Brondani et al., 2014, African-American population McKean-Cowdin et al., 2007, and the Tatar population Kochetova et al., 2015. But they are contrary to findings in other studies Baturin et al., 2014Fujisawa et al., 1998Kurokawa et al., 2001Szendrei et al., 2016. This inconsistency from findings of other studies could be explained by different genetic backgrounds of the studied populations, or to lack of randomization, small sample size, and/or presence of confounding variables such as age or sex.
No significant effects of the ADRB3/rs4994 polymorphism on blood lipid profile or blood pressure were observed, except for the CT genotype which was associated with a marginally significant increase in HDL levels (t=1.3, p= 0.09) and significantly associated with lower systolic blood pressure (t=-1.6, p= 0.05).
ADRB3 is believed to induce brown fat thermogenesis and thus resist obesity. The missense mutation of ADRB3/rs4994 results in tryptophan to arginine substitution in the first coding exon of the receptor Candelore et al., 1996. The effect of this substitution on the function of ADRB3 is not clear. Clinical and in vitro studies have suggested that this mutation reduces the sensitivity of ADRB3 to its selective and non-selective agonists Candelore et al., 1996Hoffstedt et al., 1999. In light of the current evidence, the association of wildtype rs4994 with obesity cannot be explained. It is possible that one’s ethnic background alters the effect of this mutation on obesity and fat metabolism. Further studies are required to explore the precise mechanisms behind the association.
In our study we had some limitations, such as small sample size and lack of randomization. Furthermore, the presence of confounding factors (e.g. socio-economical status, variety of eating habits, education, sex, and marital status) can all affect BMI. Caution is needed when interpreting the data of this study; further studies are required to confirm our findings.
Conclusion
In conclusion, our study suggests that the mutated allele of the ADRB3/rs4994 polymorphism may reduce the risk of obesity among Saudis. Further studies are needed to confirm our findings and to further explore the role of ADRB3/rs4994 in obesity.
Author Contribution
All the authors contribute significantly and equally to all parts of the study. All authors read and approved the final manuscript.
References
-
K. K.
Abu-Amero,
O. M.
Al-Boudari,
G. H.
Mohamed,
N.
Dzimiri.
Beta 3 adrenergic receptor Trp64Arg polymorphism and manifestation of coronary artery disease in Arabs. Human Biology.
2005;
77(6)
:
795-802
.
View Article PubMed Google Scholar -
S.
Al Dahi,
I.
Al Hariri,
W.
Al-Enazy.
Prevalence of overweight and obesity among Saudi primary school students in Tabuk, Saudi Arabia. Saudi Journal of Obesity.
2014;
2(1)
:
13
.
View Article Google Scholar -
P.
Bacchetti,
J. M.
Leung.
Sample size calculations in clinical research. Anesthesiology.
2002;
97(4)
:
1028-1029
.
View Article Google Scholar -
A. K.
Baturin,
E. Iu.
Sorokina,
A. V.
Pogozheva,
E. V.
Peskova,
O. N.
Makurina,
V. A.
Tutel’ian.
[Regional features of obesity-associated gene polymorphism (rs9939609 FTO gene and gene Trp64Arg ADRB3) in Russian population]. Voprosy Pitaniia.
2014;
83(2)
:
35-41
.
PubMed Google Scholar -
L. A.
Brondani,
G. C. K.
Duarte,
L. H.
Canani,
D.
Crispim.
The presence of at least three alleles of the ADRB3 Trp64Arg (C/T) and UCP1 -3826A/G polymorphisms is associated with protection to overweight/obesity and with higher high-density lipoprotein cholesterol levels in Caucasian-Brazilian patients with type 2 diabetes. Metabolic Syndrome and Related Disorders.
2014;
12(1)
:
16-24
.
View Article PubMed Google Scholar -
M. R.
Candelore,
L.
Deng,
L. M.
Tota,
L. J.
Kelly,
M. A.
Cascieri,
C. D.
Strader.
Pharmacological characterization of a recently described human beta 3-adrenergic receptor mutant. Endocrinology.
1996;
137(6)
:
2638-2641
.
View Article PubMed Google Scholar -
A. M.
Cypess,
L. S.
Weiner,
C.
Roberts-Toler,
E.
Franquet Elía,
S. H.
Kessler,
P. A.
Kahn,
G. M.
Kolodny.
Activation of human brown adipose tissue by a β3-adrenergic receptor agonist. Cell Metabolism.
2015;
21(1)
:
33-38
.
View Article PubMed Google Scholar -
D. A.
Luis,
M.
Gonzalez Sagrado,
R.
Aller,
O.
Izaola,
R.
Conde.
Influence of the Trp64Arg polymorphism in the beta 3 adrenoreceptor gene on insulin resistance, adipocytokine response, and weight loss secondary to lifestyle modification in obese patients. European Journal of Internal Medicine.
2007;
18(8)
:
587-592
.
View Article PubMed Google Scholar -
T.
Fujisawa,
H.
Ikegami,
Y.
Kawaguchi,
T.
Ogihara.
Meta-analysis of the association of Trp64Arg polymorphism of beta 3-adrenergic receptor gene with body mass index. The Journal of Clinical Endocrinology and Metabolism.
1998;
83(7)
:
2441-2444
.
View Article PubMed Google Scholar -
J.
Hoffstedt,
O.
Poirier,
A.
Thörne,
F.
Lönnqvist,
S. M.
Herrmann,
F.
Cambien,
P.
Arner.
Polymorphism of the human β3-adrenoceptor gene forms a well-conserved haplotype that is associated with moderate obesity and altered receptor function. Diabetes.
1999;
48(1)
:
203-205
.
View Article PubMed Google Scholar -
O. V.
Kochetova,
T. V.
Viktorova,
O. E.
Mustafina,
A. A.
Karpov,
E. K.
Khusnutdinova.
Genetic association of ADRA2A and ADRB3 genes with metabolic syndrome among the Tatars. Russian Journal of Genetics.
2015;
51(7)
:
830-834
.
View Article PubMed Google Scholar -
N.
Kurokawa,
K.
Nakai,
S.
Kameo,
Z. M.
Liu,
H.
Satoh.
Association of BMI with the beta3-adrenergic receptor gene polymorphism in Japanese: Meta-analysis. Obesity Research.
2001;
9(12)
:
741-745
.
View Article PubMed Google Scholar -
N.
Kurokawa,
E. H.
Young,
Y.
Oka,
H.
Satoh,
N. J.
Wareham,
M. S.
Sandhu,
R. J. F.
Loos.
The ADRB3 Trp64Arg variant and BMI: A meta-analysis of 44 833 individuals. International Journal of Obesity.
2008;
32(8)
:
1240-1249
.
View Article PubMed Google Scholar -
J.
Liu,
B.
Zhang,
M.
Li,
C.
Li,
Y.
Liu,
Z.
Wang,
S.
Wen.
[Study on relationship between Trp64Arg polymorphism of β3-adrenergic receptor gene and obesity and blood lipids]. Zhonghua Yi Xue Za Zhi.
2015;
95(20)
:
1558-1562
.
PubMed Google Scholar -
R.
McKean-Cowdin,
X.
Li,
L.
Bernstein,
A.
McTiernan,
R.
Ballard-Barbash,
W. J.
Gauderman,
F.
Gilliland.
The ADRB3 Trp64Arg variant and obesity in African-American breast cancer cases. International Journal of Obesity.
2007;
31(7)
:
1110-1118
.
View Article PubMed Google Scholar -
Z. A.
Memish,
C.
El Bcheraoui,
M.
Tuffaha,
M.
Robinson,
F.
Daoud,
S.
Jaber,
A. A.
Al Rabeeah.
Obesity and associated factors—Kingdom of Saudi Arabia, 2013. Preventing Chronic Disease.
2014;
11
:
E174
.
View Article PubMed Google Scholar -
A. E.
Mirrakhimov,
A. S.
Kerimkulova,
O. S.
Lunegova,
C. B.
Moldokeeva,
Y. V.
Zalesskaya,
S. S.
Abilova,
E. M.
Mirrakhimov.
An association between TRP64ARG polymorphism of the B3 adrenoreceptor gene and some metabolic disturbances. Cardiovascular Diabetology.
2011;
10(1)
:
89
.
View Article PubMed Google Scholar -
S.
Morcillo,
F.
Cardona,
G.
Rojo-Martínez,
M. C.
Almaraz,
I.
Esteva,
M. S.
Ruiz-De-Adana,
F.
Soriguer.
Effect of the combination of the variants -75G/A APOA1 and Trp64Arg ADRB3 on the risk of type 2 diabetes (DM2). Horumon To Rinsho.
2008;
68(1)
:
102-107
.
View Article PubMed Google Scholar -
C.
Santiago,
J. R.
Ruiz,
A.
Buxens,
M.
Artieda,
D.
Arteta,
M.
González-Freire,
A.
Lucia.
Trp64Arg polymorphism in ADRB3 gene is associated with elite endurance performance. British Journal of Sports Medicine.
2011;
45(2)
:
147-149
.
View Article PubMed Google Scholar -
M. A.
Shaikh,
F.
Al Sharaf,
K.
Shehzad,
F.
Shoukat,
Z.
Naeem,
S.
Al Harbi,
S.
Al Motairi.
Prevalence and trends of overweight and obesity amongst Saudi school children, a study done by using three noninvasive methods (Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5003581/). International Journal of Health Sciences.
2016;
10(3)
:
381-387
.
-
K.
Shiwaku,
A.
Nogi,
E.
Anuurad,
K.
Kitajima,
B.
Enkhmaa,
K.
Shimono,
Y.
Yamane.
Difficulty in losing weight by behavioral intervention for women with Trp64Arg polymorphism of the β3-adrenergic receptor gene. International Journal of Obesity.
2003;
27(9)
:
1028-1036
.
View Article PubMed Google Scholar -
B.
Szendrei,
D.
González-Lamuño,
T.
Amigo,
G.
Wang,
Y.
Pitsiladis,
P. J.
Benito,
R.
Cupeiro,
the PRONAF Study Group..
Influence of ADRB2 Gln27Glu and ADRB3 Trp64Arg polymorphisms on body weight and body composition changes after a controlled weight-loss intervention. Applied Physiology, Nutrition, and Metabolism.
2016;
41(3)
:
307-314
.
View Article PubMed Google Scholar -
S.
Takeuchi,
T.
Katoh,
T.
Yamauchi,
Y.
Kuroda.
ADRB3 polymorphism associated with BMI gain in Japanese men. Experimental Diabetes Research.
2012;
2012
:
973561
.
View Article PubMed Google Scholar -
T.
Umekawa,
T.
Yoshida,
N.
Sakane,
A.
Kogure,
M.
Kondo,
H.
Honjyo.
Trp64Arg mutation of β3-adrenoceptor gene deteriorates lipolysis induced by β3-adrenoceptor agonist in human omental adipocytes. Diabetes.
1999;
48(1)
:
117-120
.
View Article PubMed Google Scholar -
M.
Zawodniak-Szałapska,
R.
Stawerska,
E.
Brzeziańska,
D.
Pastuszak-Lewandoska,
J.
Lukamowicz,
K.
Cypryk,
A.
Lewiński.
Association of Trp64Arg polymorphism of beta3-adrenergic receptor with insulin resistance in Polish children with obesity. Journal of Pediatric Endocrinology & Metabolism.
2008;
21(2)
:
147-154
.
View Article PubMed Google Scholar
Comments
Downloads
Article Details
Volume & Issue : Vol 5 No 3 (2018)
Page No.: 2050-2063
Published on: 2018-03-09
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 7445 times
- Download PDF downloaded - 2143 times
- View Article downloaded - 0 times
 Biomedpress
Biomedpress