Abstract
Inflammatory bowel disease (IBD) is a heterogeneous chronic idiopathic inflammatory disease that includes ulcerative colitis (UC) and Crohn's disease with uncertain etiology and pathogenesis. The prevalence of IBD has steadily increased worldwide, affecting numerous groups of people in both developed and developing countries. Current guidelines are being re-defined with the aim for patients with established IBD to achieve mucosal healing and complete morphological figures with minimal drug toxicity, to diminish the likelihood of extra-intestinal manifestations and dysplasia/ colorectal cancer, and to prevent the need for surgery. Many patients with established IBD may require conventional treatment, as well as alternative treatment, due to non-response, loss of response, or intolerance to treatment regimen. In this context, stratification of risk and management of IBD could be based on personified strategy predominantly allocated to core components of pathogenesis of the disease. MicroRNAs (miRNAs) are defined as non-coding short RNAs which are involved in various stages of the natural evolution of IBD. Recent animal and clinical studies have shown the role of single nucleotide polymorphisms (SNPs) and signature of miRNAs in susceptibility to IBD, risk of clinical and histological exacerbation, and remission. The aim of the review is to summarize the knowledge regarding use of miRNAs as biomarkers and molecular targets in IBD.
Introduction
Inflammatory bowel disease (IBD) is defined as a group of complex and heterogeneous chronic idiopathic inflammatory diseases that includes ulcerative colitis (UC) and Crohn’s disease1. The prevalence of IBD appears to be steadily growing worldwide and affecting people of all ages, sexes, and ethnicities2. It has been suggested that several environmental, genetic and immunological factors, as well as particularities of diet, smoking, and metabolic co-morbidity states (e.g. abdominal obesity, diabetes mellitus, etc.) can modify endothelial expression of pro-inflammatory cytokines and adhesion molecules, thereby playing a pivotal role in the initiation and development of IBD6, 5, 4, 3. Current guidelines are being re-defined with the aim for patients with established IBD to achieve mucosal healing and have complete morphological figures with minimal drug toxicity. The goal also is to diminish the probability of extra-intestinal manifestations and dysplasia/ colorectal cancer, while preventing the need for surgery7.
Although the main principles of diagnosing and managing IBD have been well-reported in numerous studies and clinical guidelines10, 9, 8, there is still a lack of clear understanding of the precise etiology and pathogenesis of IBD, which warrants further investigations into novel innate molecular mechanisms of the pathogenesis of the disease11. New molecular targets are needed as the basis for targeted therapies as well as for biological markers for risk stratification during treatment12. Indeed, new biotechnological drugs, predominantly those such as monoclonal antibodies (mAbs), bispecific antibodies, specific blockers of interleukin [IL]-23, and tumor necrosis factor-alpha (TNF-α), have effectively changed the treatment paradigm for IBD patients previously allocated to be refractory to conventional treatment with corticosteroids and sulfasalazine (5-ASA). Moreover, these new drugs have opened possibilities to reduce surgery rate and hospital admissions, as well as improving quality of life; however, other approaches including risk stratification, treatment efficacy and safety monitoring are still needed in combination15, 14, 13.
In this context, biological markers reflecting several stages of pathogenesis of IBD could be used to individualize risk assessment and created personalized treatment schemes. MicroRNAs (miRNAs) are noncoding RNA molecules involved in the conduction of gene expression, predominantly as negative feedback regulators16. There is large body of evidence regarding altered miRNA expression and miRNA-related single nucleotide polymorphisms (SNPs) in immunocompetent cells, intestinal cells, resident cells and tissue mononuclear cells; they are drivers for impaired Treg, Th1 and Th17 cell function, B-cell activation and blast transformation, over-expression of inflammatory genes (e.g. TNF-α, IL-1β, IL-18, IL-22, IL-23, type I and II interferons IFNβ and IFNγ, etc.) and transcription factors (e.g. nuclear factor-κB, Stat1/ Stat3, etc.), and worsening of post-transcriptional regulation of IL-10 release19, 18, 17. All these processes are crucial for IBD initiation and development. The aim of the review is to summarize the knowledge regarding use of miRNAs as biomarkers and molecular targets in IBD.
Definition of miRNA
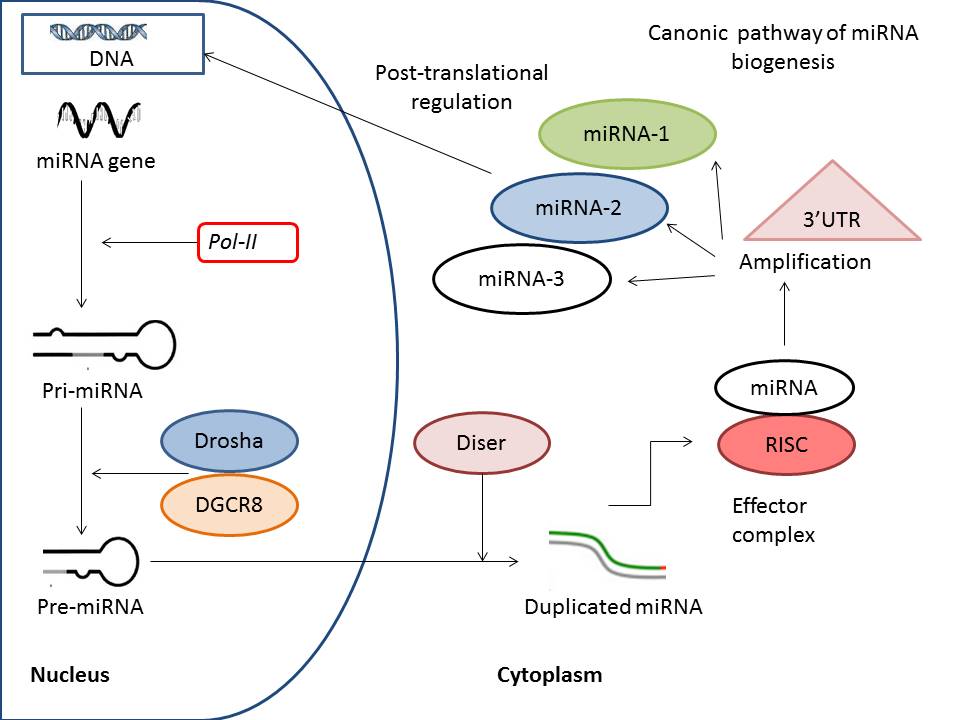
By definition, miRNAs are non-coding short RNAs about 24–30 nucleotides long, which shape RNA-protein complexes to mediate and predominantly negatively regulate epigenetic and post-transcriptional gene silencing20. According to contemporary nomeclature, miRNAs are a subtype of non-coding RNAs, which also includes other types of these molecules, such as endogenous small interfering RNAs (endo-siRNAs), Piwi-interacting RNAs (piRNAs), and long non-coding RNAs (lncRNAs)21. There are two alternative pathways to produce miRNAs: canonical and non-canonical22. The most transcribed miRNAs are a result of primary transcription of precursors of miRNA in the canonical pathway, which is characterized by consequently involving RNAse III, DROSHA, and specific microprocessor complex in cell nucleus to cleave pro-molecule and shape miRNA. The canonical biogenesis of miRNA is illustrated in Figure 1. First, in the nucleus, Drosha associated with DiGeorge syndrome critical region 8 protein shapes precursor miRNAs (pre-miRNA) from primary miRNA (pri-miRNA) transcripts. The next step in the transcription of miRNA is export of pro-molecule (pre-miRNA) into the cytoplasm with further shortening processing, which is conducted by specific RNase III enzyme Dicer. This enzymatic cascade shapes a respectively unstable, short, double-stranded molecule of miRNA that gives yield to the mature miRNA. The next step affects the incorporation of the single-stranded mature miRNA into the RNA-induced silencing complex, which is the primary target which cleaves the standing miRNA.
An alternative non-canonical biogenesis of miRNA is based on independent mechanisms including both DROSHA and DICER processes in the synthesis of miRNA. In fact, miRNAs are core post-translational regulators of cell functions through modulating activity of the nuclear transcription factor kappa-beta (NF-κB) pathway and promoting development of IBD.
Previous studies have revealed that there have been cell type-specific effects of miRNAs, while numerous miRNAs have not demonstrated tissue specificity; they have exhibited universal effects regardless of the multi-functional crosslink to metabolic responses23. The miRNAs are involved in numerous biological processes, such as immunity, cell growth, inflammation, differentiation, proliferation, and apoptosis24. Additionally, miRNAs have been discussed as a key player in tumorigenesis through remarkably promoting clonogenicity, cell migration, and invasion25. Immune function and biological roles of miRNAs in IBD are presented in Figure 2. Development of IBD have been correlated to increased circulating levels of several miRNAs (e.g. miRNA-28-5p, miRNA -103-2, miRNA -126, miRNA -151-5p, miRNA -199a-5p, miRNA -320, miRNA -340, miRNA -362-3p, and miRNA -532-3p), and numerous miRNAs have been found in lowered concentrations (e.g. miRNA-505)26. Although these findings have been revealed to be associated with IBD in the population of patients with known diagnosis, they have not explained whether signature miRNAs or SNPs of miRNAs have caused IBD.
Susceptibility for IBD and signature of miRNAs
Previous studies have shown that IBD is associated with sophisticated changes in structure and function of intestinal cells and that this process affects expression of several constitutive and inducible enzymes, including metabolizing enzymes (e.g., CYP2C9 and UGT1A1), membrane-associated co-transporters (e.g. ABCB1, ABCG2, monocarboxylate transporter 1, organic anion-transporting polypeptide 2B1, and organic cation transporter-like), as well as inflammatory cytokines (e.g. IL-33), which were all under control of microRNAs29, 28, 27. There is an assumption that altered cooperation between inflammatory-induced miRNA expression and membrane-associated regulatory molecules, enzymes and peptides has been implicated in susceptibility of IBD. Yet, SNPs of miRNAs may influence miRNA expression and/or maturation which, if as speculated, is able to translate into specific phenotypic changes.
| miRNA | SNP | Relation | Significance | Reference |
|---|---|---|---|---|
| miRNA-146a | rs2910164 | Asian population | ↓ UC risk | 18 |
| miRNA-146a | rs2910164 | Caucasian population | ↓ UC risk | 19 |
| miRNA-196a2 | rs11614913 | Asian population | ↓colorectal cancer risk | 18 |
| miRNA-196a2 | rs11614913 | Caucasian population | ↓ UC risk↓colorectal cancer risk | 19 28 |
| miRNA-196a-2 | rs11614913 | North Indian population | ↑UC risk | 29 |
| miRNA-499 | rs3746444 | North Indian population | ↑UC risk | 29 |
| miRNA-146a | rs2910164 | Chinese population | ↑UC risk | 30 |
| miRNA‑149 | rs2292832 | Chinese population | ↑UC risk | 30 |
| miRNA‑196a | rs11614913 | Chinese population | ↑ risk of transformation of UC into colorectal cancer | 30 |
Table 1 shows the clinical significance of several miRNA polymorphisms in the context of increased susceptibility to IBD and the resulting complications, including colorectal cancer. It has been found that SNP of miRNA-146 rs2910164, but not miR-196 rs11614913, was associated with lowered risk of UC in the Asian population, but not Caucasian population18. On the contrary, in the Caucasian population, both miRNA-196 rs11614913 and miRNA-146a rs2910164 polymorphisms were found to be predictors of lowered risk of UC19. Moreover, miRNA-196a2 rs11614913 was associated with a lowered risk of colorectal cancer in UC patients, whereas miRNA-146a rs2910164 did not have a similar relation28. In a study by Ranjha R et al. (2017)29, SNPs of miRNA-196a-2 rs11614913 and miRNA-499 rs3746444 were found as predictors of risk and prognosis of IBD in the North Indian population. In the Chinese population, SNPs of miRNA-146a rs2910164 and miRNA‑149 rs2292832 were found to correspond to risk for developing IBD30. Additionally, the authors reported that SNP miRNA‑196a rs11614913 indicated a progression of IBD to colorectal cancer30. Fisher, SA et al. (2008)31, using genome-wide association scan (GWAS) technique, identified several risk loci (IL-23R, IL-12B, HLA DRB1, and transcription factor genes NKX2-3 and MST1), which were common to UC and Crohn's disease, whereas autophagy genes ATG16L1 and IRGM, along with NOD2, were specific for Crohn's disease only. Therefore, the authors did not observe SNPs in miRNAs genes which corresponded to development of IBD31. In fact, SNPs of miRNAs for several pro-inflammatory genes were strongly associated with ethnicity, while the phenotyping equivalent for majority of them have not been sustainable identified. Overall, it has been suggested that the genetic predisposition for IBD could be reflected in specific pre-existing miRNA profiles.
| miRNA | Type of dysregulation | Gene target | Relation | Significance for IBD | Reference |
|---|---|---|---|---|---|
| miRNA-15 | downregulation | adenosine A2a receptor | Inflammation-associated processes | Predict gut inflammation | 43 |
| miRNA-23b | downregulation | Marcksl | Altered intercellular communication | Predict gut inflammation | 42 |
| miRNA-26b | downregulation | E3 ubiquitin ligase DIP1 | Inflammation-associated processes | discriminate between UCC and the sporadic colon cancer | 36 |
| miRNA-193a-3p | downregulation | IL-17RD | Inflammation-associated processes | Predict neoplasia and colorectal cancer | 48 |
| miR-320 | upregulation | IL-33/ST2 | Gut repair | Predict gut inflammation | 47 |
| miRNA-146 | upregulation | MyD88; NF-κB; other genes in barrier function and immune protection | Altered proportion of Th17 / Treg and B cells (CD220+) populations | Increasing susceptibility to UC | 46,39 |
| miRNA-143 / 145 | downregulation | IRS-1, K-RAS, API5, and MEK-2 | Expression of IRS-1, K-RAS, API5, and MEK-2 in target cells (HCT116 and HCA7 cells) | predispose to chronic inflammation and neoplastic progression in IBD | 45 |
| miRNA-21 | upregulation | RhoB | Reduces intestinal barrier function | Predict gut inflammation | 44,38 |
| miRNA-28-5p | downregulation | STAT | Inflammation-associated processes | Predict gut inflammation | 33 |
| miRNA -142-3p | downregulation | NOD2 | autophagy-related pathways | Predict gut inflammation | 33 |
| miRNA -122a | downregulation | Occludin | TNFα-induced gut mucosal permeability | Predict gut inflammation | 41 |
| miRNA -124 | downregulation | AHR | Reduces intestinal barrier function | Predict gut inflammation | 37 |
| miRNA -126 | downregulation | EGFL7 | Regulating cellular adhesion, proliferation, migration and invasion | Apoptotic- and inflammatory-induced tissue injury | 33 |
| miRNA -141 | downregulation | CXCL12β | Leukocytes tissue infiltration | Predict gut inflammation | 40 |
| miRNA -146 | downregulation | MIR155MIR155HG | Impaired barrier function, regulator of inflammation and innate immune system | Predict gut inflammation | 39,32 |
| miRNA -150 | downregulation | C-Myb | Apoptotic-induced gut mucosal permeability | Predict gut inflammation | 38 |
| miRNA -214 | downregulation | PDLIM2 | Inflammatory-induced gut mucosal permeability | Predict gut inflammation | 37 |
| miRNA -151-5p | downregulation | FOXO3a | TNF-α-induced gut mucosal permeability | Predict gut inflammation | 33 |
| miRNA -155 | downregulation | FOXO3a | TNF-α-induced gut mucosal permeability | Predict gut inflammation | 35 |
| miRNA -192 | downregulation | NOD2 | autophagy-related pathways | Predict gut inflammation | 33 |
| miRNA -199a-5p | downregulation | IL-17 | Gut inflammation | Predict gut inflammation | 33 |
| miRNA -301 | downregulation | BTG1 | Gut inflammation | Inflammatory-induced colon cancer | 34 |
| miRNA -320 | downregulation | NOD2 | autophagy-related pathways | Predict gut inflammation | 33 |
| miRNA -375 | downregulation | IL-10 | Gut inflammation | Inflammatory-induced colon cancer | 32 |
miRNAs as a diagnostic and predictive tool for IBD and its complications
miRNAs are core regulators of inflammation in IBD, which suggests they mediate the process of IBD-to-colorectal cancer transformation. Taking into consideration these findings, miRNAs might have diagnostic and predictive values for patients with established diagnosis of IBD. Table 2 shows the interrelationship between miRNA expression and IBD development. Benderska, N et al. (2015)34 reported that miRNA-26b, which down-regulates E3 ubiquitin ligase DIP1 in cells of mucosa lamina of intestines, turns out to be a good biological marker for inflammation-associated processes and can be used to predict sporadic colon cancer in patients with established IBD. Pekow, J et al. (2017)35 found that miRNA-193a-3p was downregulated in UC neoplasia and attenuated malignancy in colon through up-regulation of IL-17RD. Notably, this transformation may be maintained not just by altered miRNA expression but by inflammation-associated dysbiosis in the gut lumen via co-stimulation of TLR/NFκB signaling pathway49. Meanwhile, epithelial repair and restitution are reported as crucial elements in gut mucosal healing, and antigen stimulation produced by microbiota acts as a modulator of the IL-33/ST2 axis. Thus, miRNAs are warranted for evaluation as a resolution for bowel inflammation. Indeed, epithelial-derived miR-320 has been shown to be able to promote epithelial repair through activation of IL-33/ST2 cooperation; as well, deficiency of miR-320 is considered as a marker of increased inflammatory response36. It has been revealed that miRNA-146a was over-expressed in distal colon and in ileum of IBD patients37. In fact, miRNA-146a can be found in hematopoietic cells in a microbiota-independent manner50. In animal studies, miRNA-146a sufficiently restricted the expansion of numerous T cell gut populations, such as Th17, Tregs, and Tfh cells. Consequently, an association was established between expression of barrier cells and deficiency of miRNA-146a in the intestine37. As a result, it has been suggested that miRNA-146a could be involved in the regulation of gut homeostasis besides impacting mucosal surfaces and mediating susceptibility to IBD, although this assumption is needed to be confirmed in clinical studies.
Previously, it has been reported that miRNA-143 and miRNA-145, which promote gut cell repair during stress and injury, could be feedback regulators of inflammatory-induced malignancy in IBD39, 51. Indeed, miRNA-143 and miRNA-145 were down-regulated in patients with IBD and mediated expression of IRS-1, K-RAS, API5, and MEK-2 in epithelial cells of gut, thereby predicting inflammation and colon cancer39. In another study, exaggerated levels of miRNA-21 in UC biopsy materials were found40; this molecule mediated intestinal barrier function by acting via target GTPase RhoB. In fact, over-expressed miRNA-21is considered as a key regulator of intestinal epithelial tight junction permeability that increases susceptibility to malignancy. Additionally, down-regulated miRNA-122a was associated with TNFα-induced gut mucosal permeability and maintained gut inflammation43. In some animal studies, over-expressed miRNA-150 and miRNA-214 found in UC biopsy materials were associated with apoptotic-induced changes in gut barrier function44, 41. Although it remains to be established whether these findings are essential for evolution of IBD in humans, there is evidence that transcription of both miRNAs-150 and -214 can be supported by IL-6-upregulated STAT3 in colon tissues44. As a result of this action, reduced levels of tensin homolog and PDZ and LIM domain 2 (PDLIM2) can result, leading to phosphorylation of AKT, switch on of NF-κB, and increased activity of IBD. Moreover, expression of miRNA-214 has been well-correlated with morphological changes in the gut of patients with established UC44. He, C et al. (2017)48 reported that miRNA-301A could be a promoter of gut inflammation and UC-associated malignancy, resulting in the inhibition of BTG1. Other miRNAs that are potential candidates as inflammatory response promoters (as confirmed in clinical studies) are miRNA-14145, miRNA-14638, miRNA-15547, miRNA-12444, and miRNA-23b33. Zhang & Li (2018)32 have shown that miRNA-15, which is expressed in colonic tissues of IBD patients, inversely affected the expression of adenosine A2a receptor and also mediated NF-κB cascade which supports inflammation. However, there have been numerous miRNAs up-and down-regulated in IBD and whose diagnostic and predictive roles in UC and Crohn’s disease are uncertain. Thus, further investigations are required to scrutinize the complete signature of over-expressed miRNAs.
Paraskevi, A et al. (2012)42 have investigated a wide range of miRNAs and found that miRNA-16, miRNA-21, miRNA-28-5p, miRNA-151-5p, and miRNA-199a-5p were significantly overexpressed in UC and Crohn’s disease patients compared to healthy volunteers. However, miRNA-155 was most highly expressed in UC, but not in Crohn’s disease. Moreover, miRNA-155 was a potential candidate as biomarker to distinguish UC and Crohn’s disease46. On the contrary, Schaefer JS et al. (2015)52 reported on another aberrant miRNA expression profile which indicated IBD. The authors investigated a panel of 89 miRNAs and found a signature of miRNAs which consisted of miRNA-19a, miRNA-21, miRNA-31, miRNA-101, miRNA-146a, and miRNA-375; this signature could serve as biomarkers to discriminate between UC and Crohn’s disease. Viennois, E et al. (2017)53 revealed that the signature of miRNAs (up-regulated miRNA-29b-3p, miRNA -122-5p, miRNA -192-5p, miRNA -194-5p, miRNA -375-3p, miRNA -150-5p, miRNA -146a-3p, and down-regulated miRNA-148a-3p and miRNA -199a-3p) had a high discriminatory ability for IBD, associated with Disease Activity Index, and exhibited good specificity for the presence of intestinal inflammation due to Crohn’s disease. Identification of deferentially expressed miRNAs affecting autophagy activity, pro-inflammatory cytokine production, and gut repair suggests that this miRNA signature profile could serve as a new diagnostic tool for different types of IBD. Thus, numerous small miRNAs that are up- or down-regulated in IBD can be important in IBD. However, miRNA-155 appears to be a good biomarker for distinguishing UC from Crohn’s disease. Perhaps, the determination of a signature (profile) for circulating or expressed miRNAs could be the most reliable diagnostic and predictive tool for IBD.
miRNA signature as predictor of drug response in IBD
In fact, in patients with IBD, clinical and histologic remissions do not accompany each other. There is evidence that impaired miRNA profile could be associated with clinical and histologic remission in patients treated with xenobiotic and biologically active anti-inflammatory drugs. Indeed, targeted treatment of UC and Crohn’s disease appears to be crucial to identify possible modalities of further action for the drugs, including assays to assess efficacy and toxicity. Minacapelli, CD et al. (2019)53 found that downregulated levels of miRNA-206 during clinically effective long-term mesalamine treatment of UC were associated with maintenance of anti-inflammatory A3 adenosine receptor expression in epithelial cells of the gut53. The authors concluded that miRNA-206 expression level can be used as a biological marker for prediction of positive therapeutic response to mesalamine treatment in UC. Perhaps, monitoring the immune status in IBD might be necessary when designing treatment regimens based on combination of conventional xenobiotic drugs (corticosteroids, mesalamine, etc.) and biological active drugs, including anti-TNF-α Abs/ blockers of soluble receptors for TNF-α, anti-IL-6 Abs, anti-IL-2 Abs, and anti-IL-8 Abs55, 54. Additionally, an attractive alternative to control UC could be the choice of optimal treatment taking into consideration the personalized response of each patient with IBD after the initial drug is given. Notably, this approach could be especially intriguing in non-naïve patients treated with TNF-α Abs /antagonists and in IBD patients who are candidates for biological therapies other than TNF-α Abs (ustekinumab, vedolizumab and tofacitinib)56. In this context, the miRNA signature is, indeed, a great predictor of response towards treatment and a rational approach for designing personalized treatments57. However, there is still limited evidence regarding these assumptions and, therefore, large clinical trials are required in the future to better discern whether biomarker-guided therapy of IBD could be an effective approach for the clinical treatment of IBD as hoped.
Conclusions
In conclusion, miRNA profiles are dysregulated in patients at risk for IBD and with established diagnosis of UC and Crohn’s disease. Determining an altered signature of miRNAs appears to be more preferable over single SNPs or up- and down-regulated RNAs for determining risk of susceptibility for IBD, diagnosis of the disease, transformation of UC/Crohn’s disease into colon cancer, as well as predictive response to initial therapy. More clinical trials are also needed to evaluate the role of altered immune phenotype of miRNAs in IBD and in personalized treatment of IBD.
Abbreviations
3′ UTR: 3′ untranslated region
DGCR 8: DiGeorge syndrome critical region 8 protein
DIP1: ubiquitin protein ligase 1
GWAS: genome-wide association scans
IBD: inflammatory bowel disease
IL: interleukin
mAbs: monoclonal antibodies
mRNA: micro ribonucleic acid
MyD8: Myeloid differentiation primary response 88 protein
NF-kB: nuclear factor kappa beta
RISC: RNA-silencing complex
SNPs: single-nucleotide polymorphisms
Stat: Signal transducer and activator of transcription
TNF-α: tumor necrosis factor α
UC: ulcerative colitis
UCC: associated colorectal carcinoma
Competing Interests
Authors have no competing interests.
Authors' Contributions
Alexander E Berezin: idea of the paper, selection and analysis of the findings, preparing manuscript, review and approvel of the paper.
References
-
Sairenji
T.,
Collins
K.L.,
Evans
D.V.,
An Update on Inflammatory Bowel Disease. Primary Care.
2017;
44
(4)
:
673-92
.
View Article PubMed Google Scholar -
Maxwell
E.C.,
Grossman
A.B.,
Advances in Pediatric Inflammatory Bowel Disease. Adolescent Medicine: State of the Art Reviews.
2016;
27
(1)
:
57-74
.
PubMed Google Scholar -
Hart
A.,
Diet in the etiology of inflammatory bowel disease. Revista Espanola de Enfermedades Digestivas.
2019;
111
(1)
:
3-4
.
View Article PubMed Google Scholar -
Jin
Z.Q.,
Lu
H.G.,
Wu
Q.B.,
Ge
H.X.,
Zhou
T.T.,
Zhang
X.Q.,
A meta-analysis of dietary carbohydrate intake and inflammatory bowel disease risk: evidence from 15 epidemiology studies. Revista Espanola de Enfermedades Digestivas.
2019;
111
(1)
:
5-9
.
View Article PubMed Google Scholar -
Shrestha
M.P.,
Taleban
S.,
Obesity Is Associated with Increased Risk of Colectomy in Inflammatory Bowel Disease Patients Hospitalized with Clostridium difficile Infection. Digestive Diseases and Sciences.
2018;
64
(6)
:
1632-1639
.
View Article PubMed Google Scholar -
Niriella
M.A.,
Liyanage
I.K.,
Kodisinghe
S.K.,
Silva
A.P.,
Rajapakshe
N.,
Nanayakkara
S.D.,
Genetic associations of inflammatory bowel disease in a South Asian population. World Journal of Clinical Cases.
2018;
6
(15)
:
908-15
.
View Article PubMed Google Scholar -
Kredel
L.I.,
Schneidereit
O.,
Hoffmann
J.C.,
Siegmund
B.,
Prei\ss
J.C.,
Guideline recommendations for treatment of patients with inflammatory bowel diseases are not implemented in clinical practice-results of a non-representative survey. International Journal of Colorectal Disease.
2018;
34
(3)
:
431-440
.
View Article PubMed Google Scholar -
Danese
S.,
Banerjee
R.,
Cummings
J.F.,
Dotan
I.,
Kotze
P.G.,
Leong
R.W.,
Consensus recommendations for patient-centered therapy in mild-to-moderate ulcerative colitis: the i Support Therapy-Access to Rapid Treatment (iSTART) approach. Intestinal Research.
2018;
16
(4)
:
522-8
.
View Article PubMed Google Scholar -
Magro
F.,
Gionchetti
P.,
Eliakim
R.,
Ardizzone
S.,
Armuzzi
A.,
Acosta
M. Barreiro-de,
Third European evidencebased consensus on diagnosis and management of ulcerative colitis. Part 1: definitions, diagnosis, extra-intestinal manifestations, pregnancy, cancer surveillance, surgery, and ileo-anal pouch disorders. Journal of Crohn's and Colitis.
2017;
11
(6)
:
649-70
.
View Article PubMed Google Scholar -
Kornbluth
A.,
Sachar
D.B.,
Practice Parameters Committee of the American College of Gastroenterology
Ulcerative colitis practice guidelines in adults: American College Of Gastroenterology, Practice Parameters Committee. The American Journal of Gastroenterology.
2010;
105
(3)
:
501-23
.
View Article PubMed Google Scholar -
Fiorino
G.,
Allocca
M.,
Chaparro
M.,
Coenen
S.,
Fidalgo
C.,
Younge
L.,
Quality of Care' Standards in Inflammatory Bowel Disease: A Systematic Review. Journal of Crohn's and Colitis.
2018;
13
(1)
:
127-137
.
View Article PubMed Google Scholar -
Targownik
L.E.,
Benchimol
E.I.,
Bernstein
C.N.,
Singh
H.,
Lix
M.L.,
Tennakoon
A.,
Upfront Combination Therapy, Compared With Monotherapy, for Patients Not Previously Treated With a Biologic Agent Associates With Reduced Risk of Inflammatory Bowel Disease-related Complications in a Population-based Cohort Study. Clinical Gastroenterology and Hepatology.
2018;
17
(9)
:
1788 -1798
.
View Article Google Scholar -
Podolsky
D.K.,
Inflammatory bowel disease. The New England Journal of Medicine.
2002;
347
(6)
:
417-29
.
View Article PubMed Google Scholar -
Macaluso
F.S.,
Orlando
A.,
Cottone
M.,
Anti-interleukin-12 and anti-interleukin-23 agents in Crohn's disease. Expert Opinion on Biological Therapy.
2018;
\textbullet\textbullet\textbullet
.
View Article PubMed Google Scholar -
Peyrin-Biroulet
L.,
Demarest
S.,
Nirula
A.,
The next generation of targeted inflammatory bowel disease therapies. Autoimmun Rev.
2019;
18
(2)
:
123-128
.
View Article Google Scholar -
Moein
S.,
Vaghari-Tabari
M.,
Qujeq
D.,
Majidinia
M.,
Nabavi
S.M.,
Yousefi
B.,
MiRNAs and inflammatory bowel disease: an interesting new story. Journal of Cellular Physiology.
2018;
234
(4)
:
3277-3293
.
View Article PubMed Google Scholar -
Sanctuary
M.R.,
Huang
R.H.,
Jones
A.A.,
Luck
M.E.,
Aherne
C.M.,
Jedlicka
P.,
miR-106a deficiency attenuates inflammation in murine IBD models. Mucosal Immunology.
2019;
12
(1)
:
200-11
.
View Article PubMed Google Scholar -
Li
Z.,
Wang
Y.,
Zhu
Y.,
Association of miRNA-146a rs2910164 and miRNA-196 rs11614913 polymorphisms in patients with ulcerative colitis: A meta-analysis and review. Medicine.
2018;
97
(39)
:
e12294
.
View Article PubMed Google Scholar -
Erdmann
P.,
Bruckmueller
H.,
Martin
P.,
Busch
D.,
Haenisch
S.,
Müller
J.,
Dysregulation of Mucosal Membrane Transporters and Drug-Metabolizing Enzymes in Ulcerative Colitis. J Pharm Sci.
2018;
108
(2)
:
1035-1046
.
View Article PubMed Google Scholar -
Desvignes
T.,
Batzel
P.,
Berezikov
E.,
Eilbeck
K.,
Eppig
J.T.,
McAndrews
M.S.,
miRNA Nomenclature: A View Incorporating Genetic Origins, Biosynthetic Pathways, and Sequence Variants. Trends in Genetics.
2015;
31
(11)
:
613-26
.
View Article PubMed Google Scholar -
Kim
V.N.,
Han
J.,
Siomi
M.C.,
Biogenesis of small RNAs in animals. Nature Reviews. Molecular Cell Biology.
2009;
10
(2)
:
126-39
.
View Article PubMed Google Scholar -
Castellano
L.,
Stebbing
J.,
Deep sequencing of small RNAs identifies canonical and non-canonical miRNA and endogenous siRNAs in mammalian somatic tissues. Nucleic Acids Research.
2013;
41
(5)
:
3339-51
.
View Article PubMed Google Scholar -
He
L.,
Hannon
G.J.,
MicroRNAs: small RNAs with a big role in gene regulation. Nature Reviews. Genetics.
2004;
5
(7)
:
522-31
.
View Article PubMed Google Scholar -
Matsumoto
A.,
Nakayama
K.I.,
Hidden Peptides Encoded by Putative Noncoding RNAs. Cell Structure and Function.
2018;
43
(1)
:
75-83
.
View Article PubMed Google Scholar -
Schetter
A.J.,
Heegaard
N.H.,
Harris
C.C.,
Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis.
2010;
31
(1)
:
37-49
.
View Article PubMed Google Scholar -
Xu
X.M.,
Zhang
H.J.,
miRNAs as new molecular insights into inflammatory bowel disease: crucial regulators in autoimmunity and inflammation. World Journal of Gastroenterology.
2016;
22
(7)
:
2206-18
.
View Article PubMed Google Scholar -
Annese
V.,
Piepoli
A.,
Latiano
A.,
Lombardi
G.,
Napolitano
G.,
Caruso
N.,
HLA-DRB1 alleles may influence disease phenotype in patients with inflammatory bowel disease: a critical reappraisal with review of the literature. Diseases of the Colon and Rectum.
2005;
48
(1)
:
57-64
.
View Article PubMed Google Scholar -
Wan
D.,
Gu
W.,
Xu
G.,
Shen
C.,
Ding
D.,
Shen
S.,
Effects of common polymorphisms rs2910164 in miR-146a and rs11614913 in miR-196a2 on susceptibility to colorectal cancer: a systematic review meta-analysis. Clin Transl Oncol.
2014;
16
(9)
:
792-800
.
View Article PubMed Google Scholar -
Ranjha
R.,
Meena
N.K.,
Singh
A.,
Ahuja
V.,
Paul
J.,
Association of miR-196a-2 and miR-499 variants with ulcerative colitis and their correlation with expression of respective miRNAs. PLoS One.
2017;
12
(3)
:
e0173447
.
View Article PubMed Google Scholar -
Zhu
M.,
Li
D.,
Jin
M.,
Li
M.,
Association between microRNA polymorphisms and the risk of inflammatory bowel disease. Molecular Medicine Reports.
2016;
13
(6)
:
5297-308
.
View Article PubMed Google Scholar -
Fisher
S.A.,
Tremelling
M.,
Anderson
C.A.,
Gwilliam
R.,
Bumpstead
S.,
Prescott
N.J.,
Wellcome Trust Case Control Consortium
Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nature Genetics.
2008;
40
(6)
:
710-2
.
View Article PubMed Google Scholar -
Schaefer
J.S.,
Attumi
T.,
Opekun
A.R.,
Abraham
B.,
Hou
J.,
Shelby
H.,
MicroRNA signatures differentiate Crohn's disease from ulcerative colitis. BMC Immunology.
2015;
16
(1)
:
5
.
View Article PubMed Google Scholar -
Paraskevi
A.,
Theodoropoulos
G.,
Papaconstantinou
I.,
Mantzaris
G.,
Nikiteas
N.,
Gazouli
M.,
Circulating MicroRNA in inflammatory bowel disease. Journal of Crohn's and Colitis.
2012;
6
(9)
:
900-4
.
View Article PubMed Google Scholar -
He
C.,
Yu
T.,
Shi
Y.,
Ma
C.,
Yang
W.,
Fang
L.,
MicroRNA 301A Promotes Intestinal Inflammation and Colitis-Associated Cancer Development by Inhibiting BTG1. Gastroenterology.
2017;
152
(6)
.
View Article PubMed Google Scholar -
Szucs
D.,
Béres
N.J.,
Rokonay
R.,
Boros
K.,
Borka
K.,
Kiss
Z.,
Increased duodenal expression of miR-146a and -155 in pediatric Crohn's disease. World Journal of Gastroenterology.
2016;
22
(26)
:
6027-35
.
View Article PubMed Google Scholar -
Benderska
N.,
Dittrich
A.L.,
Knaup
S.,
Rau
T.T.,
Neufert
C.,
Wach
S.,
miRNA-26b Overexpression in Ulcerative Colitis-associated Carcinogenesis. Inflammatory Bowel Diseases.
2015;
21
(9)
:
2039-51
.
View Article PubMed Google Scholar -
Polytarchou
C.,
Hommes
D.W.,
Palumbo
T.,
Hatziapostolou
M.,
Koutsioumpa
M.,
Koukos
G.,
MicroRNA214 is associated with progression of ulcerative colitis, and inhibition reduces development of colitis and colitis-associated cancer in mice. Gastroenterology.
2015;
149
(4)
:
981-92.e11
.
View Article PubMed Google Scholar -
Bian
Z.,
Li
L.,
Cui
J.,
Zhang
H.,
Liu
Y.,
Zhang
C.Y.,
Role of miR-150-targeting c-Myb in colonic epithelial disruption during dextran sulphate sodium-induced murine experimental colitis and human ulcerative colitis. The Journal of Pathology.
2011;
225
(4)
:
544-53
.
View Article PubMed Google Scholar -
Wu
F.,
Zhang
S.,
Dassopoulos
T.,
Harris
M.L.,
Bayless
T.M.,
Meltzer
S.J.,
Identification of microRNAs associated with ileal and colonic Crohn's disease. Inflammatory Bowel Diseases.
2010;
16
(10)
:
1729-38
.
View Article PubMed Google Scholar -
Huang
Z.,
Shi
T.,
Zhou
Q.,
Shi
S.,
Zhao
R.,
Shi
H.,
miR-141 Regulates colonic leukocytic trafficking by targeting CXCL12β during murine colitis and human Crohn's disease. Gut.
2014;
63
(8)
:
1247-57
.
View Article PubMed Google Scholar -
Ye
D.,
Guo
S.,
Al-Sadi
R.,
Ma
T.Y.,
MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology.
2011;
141
(4)
:
1323-33
.
View Article PubMed Google Scholar -
Takagi
T.,
Naito
Y.,
Mizushima
K.,
Hirata
I.,
Yagi
N.,
Tomatsuri
N.,
Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. Journal of Gastroenterology and Hepatology.
2010;
25
:
129-33
.
View Article PubMed Google Scholar -
Zhang
H.,
Li
W.,
microRNA-15 Activates NF-κB Pathway via Down Regulating Expression of Adenosine A2 Receptor in Ulcerative Colitis. Cellular Physiology and Biochemistry.
2018;
51
(4)
:
1932-44
.
View Article PubMed Google Scholar -
Yang
Y.,
Ma
Y.,
Shi
C.,
Chen
H.,
Zhang
H.,
Chen
N.,
Overexpression of miR-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the Rho GTPase RhoB. Biochemical and Biophysical Research Communications.
2013;
434
(4)
:
746-52
.
View Article PubMed Google Scholar -
Pekow
J.R.,
Dougherty
U.,
Mustafi
R.,
Zhu
H.,
Kocherginsky
M.,
Rubin
D.T.,
miR-143 and miR-145 are downregulated in ulcerative colitis: putative regulators of inflammation and protooncogenes. Inflammatory Bowel Diseases.
2012;
18
(1)
:
94-100
.
View Article PubMed Google Scholar -
Runtsch
M.C.,
Hu
R.,
Alexander
M.,
Wallace
J.,
Kagele
D.,
Petersen
C.,
MicroRNA-146a constrains multiple parameters of intestinal immunity and increases susceptibility to DSS colitis. Oncotarget.
2015;
6
(30)
:
28556-72
.
View Article PubMed Google Scholar -
Lopetuso
L.R.,
De Salvo
C.,
Pastorelli
L.,
Rana
N.,
Senkfor
H.N.,
Petito
V.,
IL-33 promotes recovery from acute colitis by inducing miR-320 to stimulate epithelial restitution and repair. Proceedings of the National Academy of Sciences of the United States of America.
2018;
115
(40)
:
9362-70
.
View Article PubMed Google Scholar -
Pekow
J.,
Meckel
K.,
Dougherty
U.,
Huang
Y.,
Chen
X.,
Almoghrabi
A.,
miR-193a-3p is a Key Tumor Suppressor in Ulcerative Colitis-Associated Colon Cancer and Promotes Carcinogenesis through Upregulation of IL17RD. Clinical Cancer Research.
2017;
23
(17)
:
5281-91
.
View Article PubMed Google Scholar -
Rodríguez-Nogales
A.,
Algieri
F.,
Garrido-Mesa
J.,
Vezza
T.,
Utrilla
M.P.,
Chueca
N.,
Intestinal anti-inflammatory effect of the probiotic Saccharomyces boulardii in DSS-induced colitis in mice: impact on microRNAs expression and gut microbiota composition. The Journal of Nutritional Biochemistry.
2018;
61
:
129-39
.
View Article PubMed Google Scholar -
Taganov
K.D.,
Boldin
M.P.,
Chang
K.J.,
Baltimore
D.,
NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America.
2006;
103
(33)
:
12481-6
.
View Article PubMed Google Scholar -
Boldin
M.P.,
Taganov
K.D.,
Rao
D.S.,
Yang
L.,
Zhao
J.L.,
Kalwani
M.,
miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. The Journal of Experimental Medicine.
2011;
208
(6)
:
1189-201
.
View Article PubMed Google Scholar -
Viennois
E.,
Zhao
Y.,
Han
M.K.,
Xiao
B.,
Zhang
M.,
Prasad
M.,
Serum miRNA signature diagnoses and discriminates murine colitis subtypes and predicts ulcerative colitis in humans. Scientific Reports.
2017;
7
(1)
:
2520
.
View Article PubMed Google Scholar -
Minacapelli
C.D.,
Bajpai
M.,
Geng
X.,
Van Gurp
J.,
Poplin
E.,
Amenta
P.S.,
miR-206 as a Biomarker for Response to Mesalamine Treatment in Ulcerative Colitis. Inflammatory Bowel Diseases.
2019;
25
(1)
:
78-84
.
View Article PubMed Google Scholar -
Sugita
A.,
Koganei
K.,
Tatsumi
K.,
Futatsuki
R.,
Kuroki
H.,
Yamada
K.,
[Recent advances in medical and surgical treatment of ulcerative colitis]. Nippon Geka Gakkai Zasshi.
2015;
116
(2)
:
99-103
.
PubMed Google Scholar -
Matsuoka
K.,
Watanabe
M.,
Clinical development of new drugs for inflammatory bowel disease. Japanese Journal of Clinical Medicine.
2017;
75
(3)
:
488-91
.
PubMed Google Scholar -
Targownik
L.E.,
Benchimol
E.I.,
Bernstein
C.N.,
Singh
H.,
Lix
M.L.,
Tennakoon
A.,
Upfront Combination Therapy, Compared with Monotherapy, for Patients Not Previously Treated with a Biologic Agent Associates With Reduced Risk of Inflammatory Bowel Disease-related Complications in a Population-based Cohort Study. Clinical Gastroenterology and Hepatology.
2018;
17
(9)
:
1788-1798.e2
.
View Article PubMed Google Scholar -
Shen
P.,
Zhang
Z.,
Zhu
K.,
Cao
H.,
Liu
J.,
Lu
X.,
Evodiamine prevents dextran sulfate sodium-induced murine experimental colitis via the regulation of NF-κB and NLRP3 inflammasome. Biomedicine and Pharmacotherapy.
2019;
110
:
786-95
.
View Article PubMed Google Scholar
Comments
Downloads
Article Details
Volume & Issue : Vol 7 No 2 (2020)
Page No.: 3622-3632
Published on: 2020-02-29
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
- HTML viewed - 8726 times
- Download PDF downloaded - 1879 times
- View Article downloaded - 0 times
 Biomedpress
Biomedpress