Abstract
The emerging Corona virus strain (severe acute respiratory syndrome corona virus-2 (SARS-CoV-2)) harbors intricate in the development of corona virus infection (COVID-19)-induced pneumonia and subsequently ameliorates lung infection. Genome sequence and interventions reveal proximal resemblance of corona virus strain COVID-19 with severe acute respiratory syndrome (SARS), transmittable to bats, suggesting similar primary hosts in the spread of infection. However, potential rapid human-to-human transmission has caused therapeutic challenges in treating a wide range of humans suffering from corona virus all over the world. However, up to now, no direct vaccines or antiviral drugs are available to treat COVID-19. Previously designed antiviral drugs and convalescent plasma are undergoing investigations as treatment for COVID-19 infected patients. Therapeutic challenges with regards to COVID-19 have prompted scientists to develop fruitful remedies to combat the pathogen. In this review, we address the role of current ongoing therapeutic strategies, , and complex mechanisms of adaptive immune system (B and T cells) to respond to viruses. Furthermore, we illustrate the current challenges in the treatment of COVID-19, managerial strategies, and ongoing and future perspectives.
Introduction
Coronaviruses, especially severe acute respiratory synsdrome-2 (SARS-CoV-2), are minute single-stranded micron sized RNA particles1, harboring intricate capability to induce lung infection and pneumonia in humans1; they are ordered in the Coronaviridae family2. Previous corona virus outbreaks in China (Guangdong) have revealed their potential capability to induce intestinal and lung infections3. However, none of the quarantine measures were adopted in the 2002 and 2003 outbreaks4, 5. Similarly, a decade later, Asian countries experienced another deadly outbreak of middle east respiratory syndrome (MERS) virus, belonging to MERS-CoV6, 7. Thus, research community was engaged and therapeutic modalities were followed to combat MERS. With the emergence of the new deadly outbreak of novel coronavirus COVID-19, there was experience and knowledge about the meticulous capability of genome mutation of the virus8, 9. Different inculcations revealed that mortality parameters are dependent on physical health, age, immune mechanism, and gender discrimination- ranging from 0.3% to 15%10, 11.
The human body is bombarded via pathogens and responds by sophisticated mechanisms to develop immunity12, 13. Innate immune cells (e.g. macrophages and dendritic cells) and adaptive immune cells (e.g. B and T lymphocytes) work in close proximal association to produce potent neutralizing antibodies with stringent potential to neutralize viruses and develop immunity14, 15. Following invasion of pathogen-associated molecular patterns (PAMPs) and viruses, antigen-presenting cells (APCs) like macrophages and dendritic cells respond quickly, processing and presenting peptide antigens to B Lymphocytes through major histocompatibility complex (pMHC) molecules16, 17. However, the way the immune system discriminates between the types of pathogens remains poorly understood. Typically, major histocompatibility complex- 1 (pMHC-1) and pMHC-2 molecules provide efficient potential to discriminate through substantial mechanism of antigen presentation18, 19. In addition, recent evidence from COVID-19 investigations have suggested that mutational interventions interlinked with pMHC molecules could be crucial to devastate COVID-19 induced pathogenicity and suggest pMHC as a suitable candidate for drug discovery20, 21.
Considering the structural complexity features of COVID-1922, currently no promising therapeutic strategies are available. However, several antiviral drugs previously employed for MERS-CoV and SARS-CoV are ongoing as therapeutic remedies after clinically-approved applications23, 24. Remdesivir, alone or in adjacent combination with chloroquine and interferon-beta (IFN-β), has been proven promising against SARS-CoV-2, and thus has been suggested as an available promising therapeutic strategy25, 26, 27. However, recently, several groups have actively reported on the efficiency of plasma-derived monoclonal antibodies from blood of COVID-19 infection-recovered patients, and have suggested the use of such antibodies as passive immunization therapy28, 29. However, considering the aforementioned strategies, there is a dire need to develop more potent antiviral drugs to target viral proteins, nucleotides, capsids and nucleosides.
Morphologic and key genome prospective features of COVID-19
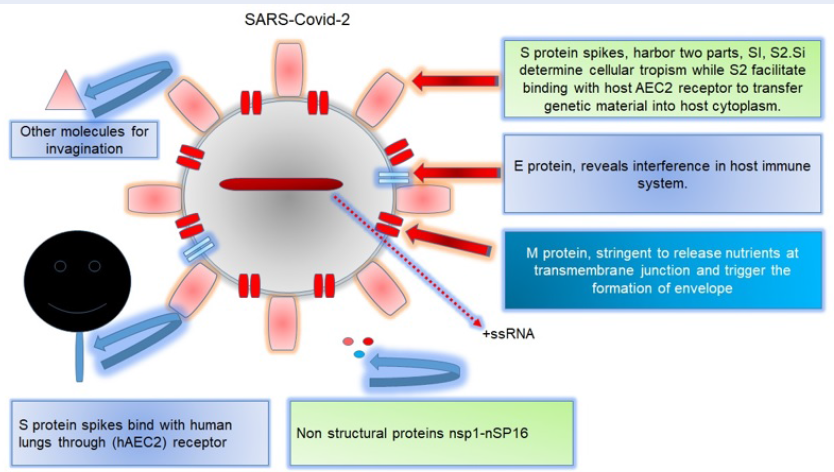
Coronaviruses - especially SARS-COVID-19 (of the Coronaviridae family) - have a morphologic configuration which shows minute single-stranded, enveloped particles ranging in size from 150-160 µm30. Further evidence have suggested that surface-flanked S proteins as well as matrix and nucleocapsid proteins harbor stringent pathogenicity factors to devastate immune mechanisms31. COVID-19 virus particles additionally encodes for hemagglutinin (HA) proteins, revealing key genome differences with other strains10, 24. Considering sequence interventions and key genome identity features, COVID-19 has been revealed to show proximal resemblance with SARS-CoV, in contrast to MERS-COVID10. In addition, surface-anchored glycoproteins and S proteins grant discriminating potential, thereby suggesting them as suitable therapeutic candidates to target surface-flanked S components. In addition, amino acid sequence interventions have revealed that S protein is divided into two parts: S1 (which facilitates COVID-19 entry into the human body), and S2 (which interferes with the host immune system)24. Similarly, additional evidence have suggested that COVID-19 shows resemblance to SARS-COVID in terms of S1, i.e. with respect to amino acid sequence interventions32. However, considering the key genome identity features, it is interesting and noteworthy that both strains gain access to invade the hosts33. Structural and conformational analyses have already revealed that human angiotension-2 (hACE-2) receptor provides an intricate attachment site to trigger COVID-19 while facilitating SARS-COVID entry into the host (Figure 1)34.
Mechanisms of the immune system in response to virus particles invading the body and the associated immunopathology
The human immune system, i.e. innate and adaptive immunity, works in close proximal coordination to produce potent neutralizing antibodies to combat virus particles invading the body35. Aberrant host immune mechanisms could initiate the onset of immunopathology with subsequent potential to devastate immune mechanism36. However, evidence have suggested that COVID-19, through human angiotensin converting enzyme 2 (hACE2) receptor, transduces genomic material into the host and mediates pattern recognition receptors (PRRs), especially toll-like receptor (TLR)-3, TLR-7 and TLR-8, which potentially detect persistence of viral particles in the cytoplasm37, thus mediating a series of immune mechanisms. Likewise, several PRRs like melanoma differentiation gene-5 (MADG-5) and retinoic acid inducible gene 1 (RIG-1) have been identified for their ability to detect cytosolic viral pathogen interlinked molecular patterns (PAMPs), triggering a proximal series of events38, 39. However, some prescribed activities initiate signaling cascade events through proximal recruitment of signaling and adaptor proteins, including mitochondria conceived antiviral protein (MAVS), stimulant of interferon gene (STING), and IFN-β, to trigger downstream series of proximal events24. However, these activities together can further lead to the recruitment of adaptor protein MyD88, which has stringent potential to regulate transcription factors, and subsequently recruits interferon-1(IFN-α/β) molecules, and pro-inflammatory cytokine candidates as key players to combat infection40.
Cytokine bombardment against COVID-19
Following viral infection, the coordinated immune system shows an intricate capability to produce pro-inflamatory cytokines41 and induce a specific lineage of T cells, including CD8+ and CD4+ T cells, as well as induction of other danger-alarming signals to potentially fight the virus42. Following infection, and injury mechanism, the pro-inflammatory cytokine milieu recruits innate immune cells, especially macrophages and granulocytes, at virus accumulated localized periphery to combat infection43. However, collectively, these efforts recruit macrophages, thus resulting in macrophage-collecting syndrome (MCS) to devastate infected tissues44. Similar inculcations have already revealed that cytokine release syndrome (CRS) is associated with the onset of morbidity leading to devastation and severity of disease45. Interestingly, interleukin (IL)-6 has been reported as a hallmark of MERS-COV infection46. Recent investigations have suggested that elevated serum concentrations of IL-6 and other pro-inflamatory cytokines result in the onset of respiratory distress and failure. Likewise, a decrease of blood cells (lymphopenia) cannot be considered as a biomarker for COVID-19 diagnosis because of its correlation with HIN1 influenza outbreak in 200946. Thus, the elevated serum interleukin level resulting in the onset of other reactive proteins is fundamental to better deciphering proximal association in terms of disease progression. Similar interventions have suggested that C reactive protein (CRP) may serve as a prognostic factor which has a contributing pivotal role in corona virus related pathologies. In addition, cytokines induced during CRS could be targeted with high-affinity antibodies to treat COV-19 patients, and have been suggested as suitable clinical drug targets46.
Overview of ongoing therapeutic strategies against SARS-Cov-2
In spite of progressive cutting-edge technologies and substantial efforts, there is currently no direct evidence of intricate therapeutic strategies to treat corona virus (COVID-19) patients37. Following infection, several healthcare organizations initially used adjacent combination of antiviral and antibacterial drugs (interferon alpha-nebulization) to reduce viral load27. Antiviral drugs (Remdesivir), alone or in adjacent combination with antibacterial (Arbidol chloroquine salts) and antimalarial drugs, have undergone ongoing tests but have been unable to achieve complete therapeutic efficacy25, 26. In addition, Remdesivir- together with interferon-β- have been clinically proven to be effective at stopping SARS-CoV-2 replication47, 48. In contrast, in spite of recovery of Covid-19 patients, higher incidences of side effects (including mental stress, and epigastric stress together with cardiac and renal complications) have been observed in elderly patients49. Similarly, in China, several Chinese traditional medicines have been administered with antiviral and antibacterial drugs to treat SARS-CoV-2 infected patients48. Plasma-isolated antibodies from blood of convalescent patients have been proven effective to neutralize COVID-19 virus, suggesting that the plasma represents an efficient and available therapeutic strategy49. Several experimental inculcations revealed the therapeutic efficacy of monoclonal antibody CR-3022 to bind with the spike glycoprotein of SARS-CoV-2, suggesting it can be an efficient therapeutic strategy in the future, possibly in combination with antibodies50. In spite of the dilemma of therapeutics, vaccines are still considered the most efficient formulation to immunize people for prevention and to treat corona virus patients51. However, in spite of progressive efforts, there are still no vaccine candidates which have been clinically approved yet to generate immune efficacy51. Following a substantial increase in SARS-CoV-2 -induced mortality, several notable pharmaceutical companies and research centers around the globe have been trying to configure a vaccine; more time is required to achieve this goal which can lead to therapeutic efficacy52. In addition, recently, PicoVac vaccine has been formulated in China and has shown some effectiveness against 10 strains of SARS-Covid-224.
Potent antiviral strategies
From phylogenetic interventions and key genome resemblance features of SARS-CoV-2 with SARS-COVID and MERS, previously designed antiviral drug Remdesivir and ribavirin have been under ongoing investigations for their potential therapeutic efficacy53. Following COVID-19 infection, some health organizations have worked together with research centers and declared adjacent treatment with chloroquine and antiviral drug Remdesivir. Interestingly, so far, Remdesivir has been a broad spectrum antiviral drug and currently tested in major countries including America, Europe and UK; it has been declared as a promising therapeutic strategy to treat SARS-CoV-2 infected patients (Table 1)26. Likewise, several broad spectrum antiviral drugs, including Ribavirin, Lopinavir, Ritonavir, Favipiravir and Umifenovir, and anti-malarial drugs (such as chloroquine and hydroxychloroquine) are being investigated in the treatment of SARS-CoV-2 infected patients34, 54, 55, 56, 57.
| Antiviral drug | Mode of activities and mechanism |
| Remdesivir | GS5734 (inhibitor of nucleoside), attained worldwide attention and frequently in use to treat COVID-19 infected patients. Remdesivir also have been employed to induce premature termination of Ebola viruses. Invivo studies revealed its potential to mitigate viral load in lungs pneumonia (Y. Cao et al., 202025; Y. Wang et al., 20204). |
| Umifenovir | Non-nucleoside immune potentiating antiviral drug, employed by Russia and China for the treatment of many corona virus pathologies and influenza prophylaxis. During current epidemic Chinese government recommend 200 mg dose thrice a day to treat COVID-19 patients (Costanzo et al., 202054; Lian et al., 202062). |
| Favipiravir | Favipiravir currently recommended in china against COVID-19 infection. Its clinical efficacy proved effective than ritonavir/ritonavir. In addition, Bangkok government declared its massive applications against COVID-19 in march. Invivo investigations declared its efficacy against SARS-CoV-2 and MERS on Vero cell lines (Cai et al., 202061 ; C. Chen et al., 202057). |
| Ritonavir/Liponavir | Alone or in adjacent combination with interferons have been recommended at 200mg/50mg dose rate twice a day. Lponavir and ritonavir have been previously used for the treatment of retroviruses, are recommended efficient against COVID-19 (B. Cao et al., 202060). |
| Hydroxychloroquine | Potential inhibitor of heme polymerase with potent antiviral strategies against SARS-CoV-2, infected patients have been approved clinically to achieve therapeutic efficacy. Triggered endosomal pH to block SARS-CoV-2 fusion events. In addition, chloroquine and hydroxychloroquine are used for the treatment of lupus nephritis and rheumatoid arthritis (RA). Likewise, invivo investigations have declared its antiviral and immune modulation activities (Colson et al., 202056; Ferner & Aronson, 202059; Gautret et al., 202063). |
| Ribavirin | Broad spectrum antiviral drug ribavirin Inhibit mRNA capping and synthesis of viral RNA. In addition, Investigations have proven effective against MERS, SARS, and SARS-CoV. However, proper dose evaluations require further research to maintain clinical efficacy (Elfiky, 202064; Taber et al., 1983)58. |
In addition, major investigations are ongoing to release large-scale slots for the betterment of humanity53. Specified targeted therapeutic interventions did not reveal obvious side effects, suggesting a comprehensive treatment modality. Thus, based on genome identity features, comprehensive drug modalities could be designed for proximal candidates of COVID-19, SARS-CoV-2, and MERS-COVID65. Furthermore, suitable drug candidates could be designed to target viral nucleotides, nucleosides and nucleic acids 53, 54. Moreover, structural investigations of surface glycoproteins (S) could offer deeper understanding for the development of potent antiviral drugs against COVID-19. More importantly, with respect to COVID-19, viral entry requires intricate cleavage at S1/S2 junction; thus, potent monoclonal antibodies could be targeted to stop S1 cleavage with subsequent application of inhibitors to stop S2 phase of infection24, 27, 28. Indeed, following SARS-COVID-19 infection in China, clinicians employed other antiviral drugs including Lopinavir/Ritonavir (LPV/RTV), which showed mild improvement only in patients harboring initial stage of symptoms within 12 days (Table 1)53.
Phylogenetic genome Interventions and key variations in SARS-CoV-2
Previously reported coronaviruses (e.g. SARS, MERS, etc.) show 80% genome identity features with SARS-CoV-224. Surface-flanked proteins are potentially encoded by four genes (M, N, S, and E), each with stringent capability to encode respective proteins including membrane proteins, nucleocapsids, surface proteins and envelope proteins, respectively, to mediate specified events24. Sixteen nonstructural proteins and pp1ab are encoded by large gene segment Off1 ab of SARS-CoV-266. According to sequence interventions, the phylogenetic tree genome of SARS-CoV-2 seems close to that of SARS coronaviruses 24. In addition, genomic sequence interventions to decipher the variations in genome features have been fundamental to better decipher strategies to generate more effective drug candidates. Genome variation analysis revealed that SARS-CoV-2 shows an absence of gene segments 8a and 8b, in contrast to SARS-CoV genome (Figure 2) 66. In addition, surface glycoprotein intervention reveals that SARS-CoV-2 surface proteins are a mixture of bat and other coronaviruses’ proteins67. Additionally, fluorescent studies conducted by some groups have reported that both SARS-coronaviruses and SARS-CoV-2 use the same proximity of ACE2 enzyme for attachment to lung38. Genome mutation interventions have demonstrated that N50IT mutations in spike S proteins trigger their intricate binding with host receptors (Figure 2)68.
Convalescent plasma as a promising option for the treatment of COVID-19 infection
Considering the outbreak of SARS-CoV-2 virus worldwide which potently induced COVID-19 infections, many research institutions and medical professionals around the world are still trying to seek suitable treatment options69. Infected elderly patients are receiving oxygenation, while severely infected people are receiving extracorporeal oxygenation to treat the corona virus induced disease24, 70. Moreover, convalescent plasma isolated from people who have recovered from COVID-19 infection has been proven as a promising therapeutic option70. Similar investigations have suggested that following infection with COVID-19, the patient’s body develops immunity at around 10-14 day after infection. Thus, administration of convalescent plasma should be considered effective69, 28. In addition, to reduce viremia, it is promising to administer convalescent plasma at early stage of disease28. Subsequent investigations are now focused on whether concurrent administration of convalescent plasma with other antiviral drugs may increase therapeutic efficacy. Similar inculcations have already revealed that co-administration of steroids, convalescent plasma, and oxygen therapy may reduce the production of antiviral antibodies28. Studies have demonstrated that frequent use of anti-steroids should be prohibited, while convalescent plasma should be tested to determine if efficacy is improved69, 70, 71.
Recent challenges in the treatment against SARS-COV-2
In spite of substantial progress in the treatment modalities against SARS-CoV-2, there are still many countries that are experiencing challenges and concerns about therapies53. Some experimental inculcations have revealed the persistence of SARS-CoV-2 in stool samples of infected patients. Detailed investigations are needed to better decipher if fecal/oral route could trigger dissemination of SARS-CoV-2 as this remains unclear72. Likewise, experimental investigations during previous SARS-CoV and MERS-CoV outbreaks have shown that the virus can survive over inanimate objects and environments for a long period; further detailed investigations into SARS-CoV-2 persistence in the environment is greatly warranted72.
Moreover, it remains unclear how efficient disinfectants are at reducing the risk of being infected with SARS-CoV-2; this area of study still requires attention and investigation73. Likewise, some experimental interventions have revealed the efficacy of Remdesivir alone to treat corona viruses; however, some cases have reported that co-administration of Remdesivir with chloroquine is effective 27, 54. Thus, detailed experimental investigations are required to solve this ambiguity. Most importantly, several studies revealed that 43% of patients suffered from fever and 15.7% suffered from pneumonia; detailed clarification and epidemiological investigations are needed to address asymptomatic carriers74. Likewise, subsequent studies are needed to decipher the timely updates in therapeutic interventions, particularly in the context of cytokine milieu, which is lacking and requires collective attention46.
Ongoing strategies used in the COVID-19 epidemic especially in Pakistan
Since there is no vaccine available against COVID-19, the prevention against the disease will remain the focus of strategy75. The primary goal should be to slow down the transmission of the virus in order to reduce the associated illnesses and deaths55. Likewise, potential ways that government and health authorities should be working in close and coordinated proximity to control the SARS-CoV-2 pandemic is a matter of utmost concern and still requires strict implementation of laws. To mitigate the mortality ratio, people in developing countries, especially, could avoid negligence and irresponsiveness when following strict strategies implemented by healthcare authorities. Thus, this prevents the chance for rapid human-human transmission and spread in communities. Therefore, each government must work together with the relevant health authorities to implement strict measures, including strict adherence to SOPs and core public health strategies, in order to control person-to-person transmission. Such strategies include identification, quarantine measures, strict individual testing, and clinical care for all cases76.
Furthermore, tracing and quarantine of all contacts should be a part of all national COVID-19 responses77. Such strategy has been successfully demonstrated in places like Wuhan, China, and has been practiced in Pakistan, as well as recognized and implemented by other countries like Germany, Vietnam and India. The actions taken by the respective governments include a range of measures in the field of public health as well as social measures to contain the spread of virus. The measures include limiting person-to-person interaction, enforcing physical and social distancing, and restricting movement. Since the emergence of the first case, governments have been effectively containing the virus. The containment measures include engaging communities both at individual and societal levels, providing information on how to protect oneself and others, interpreting scientific information into simplified messages, and encouraging the sharing of information at individual levels; all of these are fundamental to stop the risk of spreading.
Conclusion
In this review, we summarized the ongoing therapeutic interventions to treat SARS-CoV-2, key morphological features, genome variations, immunogenomics, and current challenges in treatment. Furthermore, we illustrated the current ongoing managerial strategies to manage the SARS-CoV-2 pandemic, especially in Pakistan. These revelations could lend future support for the discovering of more effective therapeutic modalities for COVID-19.
Abbreviations
APCs: antigen presenting cells
LPV, RTV: Liponavir / Ritonavir
MADG-5: melanoma differentiation gene-5
MAVS: mitochondria conceived antiviral protein
MCS: macrophage collecting syndrome
MERS: middle east respiratory syndrome
PAMPs: pathogen associated molecular patterns.
RIG-1: retinoic acid inducible gene 1
SARS-CoV: Severe acute respiratory syndrome
SARS-CoV-2: severe acute respiratory syndrome corona virus-2
Acknowledgments
The authors acknowledge the institute of Microbiology, and department of pathology, University of agriculture Faisalabad, Pakistan.
Author’s contributions
Kabeer Haneef (K.H), Muhammad Umer Asghar (M.U.A) and Ashiq Ali (A.A) are the leading authors. All these authors made substantial contributions to conception, design, acquisition of data, analysis and interpretation of data; actively contribute in drafting the article and critically provide final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Anand K, Palm GJ, Mesters JR, Siddell SG, Ziebuhr J, Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J [Internet].
2002;
21
(13)
:
3213-3224
.
View Article PubMed Google Scholar -
Siddell SG, Anderson R, Cavanagh D, Fujiwara K, Klenk HD, Macnaughton MR, et al. Coronaviridae 1.. 1983;
189
:
181-189
.
View Article PubMed Google Scholar -
Zhong NS, Zheng BJ, Li YM, Poon LLM, Xie ZH, Chan KH, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet [Internet].
2003;
362
(9393)
:
1353-1358
.
-
Yang Y, Peng F, Wang R, Guan K, Jiang T, Xu G, et al. The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun [Internet].
2020;
109
:
102434
.
PubMed Google Scholar -
Ashour HM, Elkhatib WF, Rahman MM, Elshabrawy HA. Insights into the recent 2019 novel coronavirus (Sars-coV-2) in light of past human coronavirus outbreaks. Pathogens.
2020;
9
(3)
:
1-15
.
View Article PubMed Google Scholar -
Assiri A, McGeer A, Perl TM, Price CS, Al Rabeeah AA, Cummings DAT, et al. Hospital outbreak of middle east respiratory syndrome coronavirus. N Engl J Med.
2013;
369
(5)
:
407-416
.
View Article PubMed Google Scholar -
Lee J. Better understanding on MERS Corona virus outbreak in Korea. J Korean Med Sci.
2015;
30
(7)
:
835-836
.
View Article PubMed Google Scholar -
Kim Y, Cheon S, Min CK, Sohn KM, Kang YJ, Cha YJ, et al. Spread of Mutant Middle East Respiratory Syndrome Coronavirus with Reduced Affinity to Human CD26 during the South Korean Outbreak. Buchmeier MJ, editor. MBio [Internet].
2016 ;
7
(2)
:
e00019-e00016
.
View Article PubMed Google Scholar -
Hemida MG, Chu DKW, Poon LLM, Perera RAPM, Alhammadi MA, Ng HY, et al. Mers coronavirus in dromedary camel herd, Saudi Arabia. Emerg Infect Dis.
2014;
20
(7)
:
1231-1234
.
View Article PubMed Google Scholar -
Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pacific J Allergy Immunol.
2020;
38
(1)
:
1-9
.
-
Pera A, Campos C, López N, Hassouneh F, Alonso C, Tarazona R, et al. Immunosenescence: Implications for response to infection and vaccination in older people. Maturitas [Internet].
2015;
82
(1)
:
50-55
.
View Article Google Scholar -
Liuzzi AR, McLaren JE, Price DA, Eberl M. Early innate responses to pathogens: pattern recognition by unconventional human T-cells. Curr Opin Immunol [Internet].
2015;
36
:
31-37
.
View Article PubMed Google Scholar -
Finlay BB, McFadden G. Anti-Immunology: Evasion of the Host Immune System by Bacterial and Viral Pathogens. Cell [Internet].
2006;
124
(4)
:
767-782
.
View Article PubMed Google Scholar -
Flajnik MF, Pasquier L. Evolution of innate and adaptive immunity: can we draw a line?. Trends Immunol [Internet].
2004;
25
(12)
:
640-644
.
View Article PubMed Google Scholar -
Iwasaki A, Medzhitov R. Regulation of Adaptive Immunity by the Innate Immune System. Science (80- ) [Internet].
;
327
(5963)
:
291
.
View Article PubMed Google Scholar -
Heesters BA, Poel CE, Das A, Carroll MC. Antigen Presentation to B Cells. Trends Immunol [Internet].
2016;
;37
(12)
:
844-854
.
View Article PubMed Google Scholar -
Batista FD, Harwood NE. The who, how and where of antigen presentation to B cells. Nat Rev Immunol [Internet].
2009;
9
(1)
:
15-27
.
View Article PubMed Google Scholar -
Stroynowski I. Molecules Related to Class-I Major Histocompatibility Complex Antigens. Annu Rev Immunol.
1990;
8
(1)
:
501-530
.
View Article PubMed Google Scholar -
P.J.
Bjorkman,
P.
Parham,
Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem.
1990;
1990
(59)
:
253-288
.
View Article Google Scholar -
Yewdell JW, Hill AB. Viral interference with antigen presentation. Nat Immunol [Internet].
2002;
3
(11)
:
1019-1025
.
View Article PubMed Google Scholar -
Schuren ABC, Costa AI, Wiertz EJHJ. Recent advances in viral evasion of the MHC Class I processing pathway. Curr Opin Immunol [Internet].
2016;
40
:
43-50
.
View Article PubMed Google Scholar -
Tahir QM, Alqahtani SM, Alamri MA, Chen LL. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal [Internet].
2020
.
View Article Google Scholar -
Sallard E, Lescure FX, Yazdanpanah Y, Mentre F, Peiffer-Smadja N. Type 1 interferons as a potential treatment against COVID-19. Antiviral Res [Internet].
2020;
178
:
104791
.
View Article PubMed Google Scholar -
Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak- A n update on the status. Mil Med Res.
2020;
7
(1)
:
1-10
.
View Article PubMed Google Scholar -
Cao Y, Deng Q, Dai S. Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19: An evaluation of the evidence. Travel Med Infect Dis [Internet].
2020;
:
101647
.
View Article PubMed Google Scholar -
Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet [Internet].
2020;
;395
(10236)
:
1569-1578
.
-
Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res.
2020;
30
(3)
:
269-271
.
View Article PubMed Google Scholar -
Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A.
2020;
117
(17)
:
9490-9496
.
View Article PubMed Google Scholar -
Tanne JH. Covid-19: FDA approves use of convalescent plasma to treat critically ill patients. BMJ.
2020;
368
:
m1256
.
View Article PubMed Google Scholar -
Wu JT, Leung K, Bushman M, Kishore N, Niehus R, Salazar PM, et al. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med [Internet].
2020;
26
(4)
:
506-510
.
View Article PubMed Google Scholar -
Kannan S, Shaik Syed Ali P, Sheeza A, Hemalatha K. COVID-19 (Novel Coronavirus 2019) - recent trends. Eur Rev Med Pharmacol Sci.
2020;
24
(4)
:
2006-2011
.
-
Mohan C, Kumar V, Tokas T, Singh T, Saini M, Singh D. Evaluation of SARS & COVID-19 Pandemic with Prevention and Hygiene Action Plan. 2020;
7
:
228-231
.
-
Sohrabi C, Alsafi Z, O'Neill N, Khan M, Kerwan A, Al-Jabir A, et al. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int J Surg [Internet].
2020;
76
:
71-76
.
View Article PubMed Google Scholar -
Sun J, He WT, Wang L, Lai A, Ji X, Zhai X, et al. COVID-19: Epidemiology, Evolution, and Cross-Disciplinary Perspectives. Trends Mol Med [Internet].
2020;
26
(5)
:
483-495
.
View Article PubMed Google Scholar -
Koyama S, Ishii KJ, Coban C, Akira S. Innate immune response to viral infection. Cytokine [Internet].
2008;
43
(3)
:
336-341
.
View Article PubMed Google Scholar -
Bergmann CC, Yao Q, Ho CK, Buckwold SL. Flanking residues alter antigenicity and immunogenicity of multi-unit CTL epitopes. J Immunol [Internet].
1996 ;
157
(8)
:
3242 -3249
.
-
Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal [Internet].
2020;
10
(2)
:
102-108
.
PubMed Google Scholar -
Thankam FG, Agrawal DK. Molecular chronicles of cytokine burst in COVID-19 patients with cardiovascular diseases. J Thorac Cardiovasc Surg [Internet].
2020
.
View Article Google Scholar -
Flür K, Allam R, Zecher D, Kulkarni OP, Lichtnekert J, Schwarz M, et al. Viral RNA Induces Type I Interferon-Dependent Cytokine Release and Cell Death in Mesangial Cells via Melanoma-Differentiation-Associated Gene-5: Implications for Viral Infection-Associated Glomerulonephritis. Am J Pathol [Internet].
2009;
175
(5)
:
2014-2022
.
PubMed Google Scholar -
Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol [Internet].
2020;
20
(6)
:
355-362
.
View Article PubMed Google Scholar -
Mogensen TH, Paludan SR. Molecular Pathways in Virus-Induced Cytokine Production. Microbiol Mol Biol Rev.
2001;
65
(1)
:
131-150
.
View Article PubMed Google Scholar -
Marchant A, Appay V, Van Der Sande M, Dulphy N, Liesnard C, Kidd M, et al. Mature CD8+ T lymphocyte response to viral infection during fetal life. J Clin Invest.
2003;
111
(11)
:
1747-1755
.
View Article PubMed Google Scholar -
McGonagle D, Sharif K, O'Regan A, Bridgewood C. The Role of Cytokines including Interleukin-6 in COVID-19 induced Pneumonia and Macrophage Activation Syndrome-Like Disease. Autoimmun Rev [Internet].
2020;
19
(6)
:
102537
.
View Article PubMed Google Scholar -
Hui KPY, Cheung MC, Perera RAPM, Ng KC, Bui CHT, Ho JCW, et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med [Internet].
2020;
8
(7)
:
687-695
.
-
Zhang C, Wu Z, Li JW, Zhao H, Wang GQ. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int J Antimicrob Agents [Internet].
2020;
55
(5)
:
105954
.
View Article PubMed Google Scholar -
Moore JB, June CH. Cytokine release syndrome in severe COVID-19. Science (80- ) [Internet].
2020 ;
368
(6490)
:
473 -474
.
View Article PubMed Google Scholar -
Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med.
2020;
382
(10)
:
929-936
.
View Article PubMed Google Scholar -
Sheahan TP, Sims AC, Leist SR, Schäfer A, Won J, Brown AJ, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun [Internet].
2020;
11
(1)
.
View Article PubMed Google Scholar -
Zhang W, Zhao Y, Zhang F, Wang Q, Li T, Liu Z, et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin Immunol [Internet].
2020;
214
:
108393
.
View Article PubMed Google Scholar -
Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect.
2020;
9
(1)
:
382-385
.
View Article PubMed Google Scholar -
Liu C, Zhou Q, Li Y, Garner L V, Watkins SP, Carter LJ, et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent Sci [Internet].
2020;
6
(3)
:
315-331
.
View Article PubMed Google Scholar -
Cohen J. Vaccine designers take first shots at COVID-19. Science (80- ) [Internet].
2020 ;
368
(6486)
:
14-16
.
View Article PubMed Google Scholar -
Galluccio F, Ergonenc T, Garcia Martos A, Allam AES, Pérez-Herrero M, Aguilar R, et al. Treatment algorithm for COVID-19: a multidisciplinary point of view. Clin Rheumatol.
2020;
:
2077-2084
.
View Article PubMed Google Scholar -
Costanzo M, De Giglio MAR, Roviello GN. SARS CoV-2: Recent Reports on Antiviral Therapies Based on Lopinavir/Ritonavir, Darunavir/Umifenovir, Hydroxychloroquine, Remdesivir, Favipiravir and Other Drugs for the Treatment of the New Coronavirus. Curr Med Chem.
2020;
27
(00)
.
View Article PubMed Google Scholar -
Wu Z, McGoogan JM. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases from the Chinese Center for Disease Control and Prevention. JAMA - J Am Med Assoc.
2020;
323
(13)
:
1239-1242
.
View Article PubMed Google Scholar -
Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents [Internet].
2020;
55
(4)
:
105932
.
View Article PubMed Google Scholar -
Chen C, Huang J, Cheng Z, Wu J, Chen S, Zhang Y, et al. Favipiravir versus Arbidol for COVID-19: A Randomized Clinical Trial. medRxiv.
2020
.
View Article Google Scholar -
Taber
Larry H,
Knight
Vernon,
Gilbert
Brian E,
McClung
Harvey W,
Wilson
Samuel Z,
Norton
H James,
Thurson
Jeffrey M,
Gordon
Wayne H,
Atmar
Robert L,
Schlaudt
W Robin,
Ribavirin aerosol treatment of bronchiolitis associated with respiratory syncytial virus infection in infants. Pediatrics.
1983;
72
(5)
:
613-618
.
PubMed Google Scholar -
Ferner
Robin E,
Aronson
Jeffrey K,
Chloroquine and hydroxychloroquine in covid-19. BMJ .
2020;
369
:
m1432
.
View Article Google Scholar -
Cao
Bin,
Wang
Yeming,
Wen
Danning,
Liu
Wen,
Wang
Jingli,
Fan
Guohui,
Ruan
Lianguo,
Song
Bin,
Cai
Yanping,
Wei
Ming,
A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. New England Journal of Medicine.
2020;
382
(19)
:
1787-1799
.
View Article Google Scholar -
Cai
Qingxian,
Yang
Minghui,
Liu
Dongjing,
Chen
Jun,
Shu
Dan,
Xia
Junxia,
Liao
Xuejiao,
Gu
Yuanbo,
Cai
Qiue,
Yang
Yang,
Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering.
2020;
:
Article in press
.
View Article PubMed Google Scholar -
Lian
Ningfang,
Xie
Hansheng,
Lin
Su,
Huang
Jiefeng,
Zhao
Jianming,
Lin
Qichang,
Umifenovir treatment is not associated with improved outcomes in patients with coronavirus disease 2019: a retrospective study. Clinical Microbiology and Infection.
2020;
26
(2020)
:
917-921
.
View Article Google Scholar -
Gautret
Philippe,
Lagier
Jean-Christophe,
Parola
Philippe,
Meddeb
Line,
Mailhe
Morgane,
Doudier
Barbara,
Courjon
Johan,
Giordanengo
Valérie,
Vieira
Vera Esteves,
Dupont
Hervé Tissot,
Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. International journal of antimicrobial agents.
2020;
56
(1)
:
105949
.
View Article Google Scholar -
Elfiky
Abdo A,
Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life sciences.
2020;
253
(2020)
:
117592
.
View Article PubMed Google Scholar -
Ceraolo C, Giorgi FM. Genomic variance of the 2019-nCoV coronavirus. J Med Virol [Internet].
2020;
92
(5)
:
522-528
.
View Article PubMed Google Scholar -
Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe [Internet].
2020;
27
(3)
:
325-328
.
View Article PubMed Google Scholar -
Li B, Si HR, Zhu Y, Yang XL, Anderson DE, Shi ZL, et al. Discovery of Bat Coronaviruses through Surveillance and Probe Capture-Based Next-Generation Sequencing. mSphere.
2020;
5
(1)
:
1-11
.
View Article PubMed Google Scholar -
Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Gallagher T, editor. J Virol [Internet].
2020 ;
94
(7)
:
e00127-e00120
.
View Article PubMed Google Scholar -
Ye M, Fu D, Ren Y, Wang F, Wang D, Zhang F, et al. Treatment with convalescent plasma for COVID-19 patients in Wuhan, China. J Med Virol [Internet].
.
View Article PubMed Google Scholar -
Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis [Internet].
2020;
20
(4)
:
398-400
.
View Article Google Scholar -
Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci [Internet].
2020 ;
117
(17)
:
9490 -9496
.
-
Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents [Internet].
2020;
55
(3)
:
105924
.
View Article PubMed Google Scholar -
Kampf G, Todt D, Pfaender S, Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect [Internet].
2020;
104
(3)
:
246-251
.
View Article PubMed Google Scholar -
Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv [Internet].
2020
.
-
Arabi YM, Fowler R, Hayden FG. Critical care management of adults with community-acquired severe respiratory viral infection. Intensive Care Med [Internet].
2020;
46
(2)
:
315-328
.
View Article PubMed Google Scholar -
Anderson RM, Heesterbeek H, Klinkenberg D, Hollingsworth TD. How will country-based mitigation measures influence the course of the COVID-19 epidemic?. Lancet.
2020;
395
(10228)
:
931-934
.
View Article Google Scholar -
Getaneh Y, Yizengaw A, Adane S, Zealiyas K, Abate Z, Leulseged S, et al. Global lessons and Potential strategies in combating COVID-19 pandemic in Ethiopia: Systematic Review. medRxiv [Internet].
2020
.
View Article Google Scholar
Comments
Downloads
Article Details
Volume & Issue : Vol 7 No 8 (2020)
Page No.: 3906-3915
Published on: 2020-08-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 8290 times
- Download PDF downloaded - 2109 times
- View Article downloaded - 0 times
 Biomedpress
Biomedpress



