Abstract
Introduction: The emergence of a novel coronavirus, SARS-CoV-2, an etiologic agent of coronavirus disease (COVID-19), has led to a pandemic of global concern. Considering the huge number of morbidity and mortality worldwide, the World Health Organization declared, on 11th March 2020, the pandemic as an unprecedented public health crisis. The virus is a member of plus sense RNA viruses that can show a high rate of mutations. The ongoing multiple mutations in the structural proteins of coronavirus drive viral evolution, enabling them to evade the host immunity and rapidly acquire drug resistance. In the present study, we focused mainly on the prevalence of mutations in the four types of structural proteins- S (spike), E (envelope), M (membrane), and N (nucleocapsid)- that are required for the assembly of a complete virion particle. Further, we estimated the antigenicity and allergenicity of these structural proteins to design and develop a potentially good candidate vaccine against SARS-CoV-2.
Methods: In the present in silico study, envelope protein was found to be highly antigenic, followed by the nucleocapsid, membrane, and spike proteins of SARS-CoV-2.
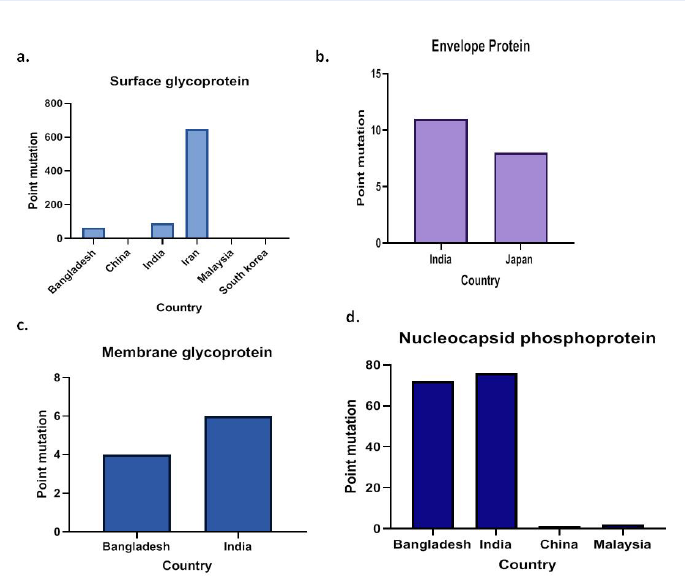
Results: In this study, we detected 987 mutations from 729 sequences from Asia in October 2020, and compared them with China's first Wuhan isolate sequence as a reference. Spike protein showed the highest mutations with 807 point mutations among the four structural proteins, followed by nucleocapsid with 151 mutations, while envelope showed 19 mutations and membrane only 10 point mutations.
Conclusion: Taken together, our study revealed that variations occurring in the structural protein of SARS-CoV-2 might be altering the viral structure and function, and that the envelope protein appears to be a promising vaccine candidate to curb coronavirus infections.
Introduction
Human Coronavirus (SARS-CoV-2, Severe acute respiratory syndrome) is a positive-sense RNA virus. As an etiologic agent of coronavirus disease 2019 (COVID-19), the virus induces moderate to severe respiratory distress1. This pandemic originated from an animal market in Wuhan city of China2. The ripple effect of this contagious viral disease has created a humanitarian health crisis and has become an enormous challenge to the entire health systems across the globe.
SARS-CoV-2 is a member of the Coronaviridae family and Nidovirales order. The virus is considered the third zoonotic coronavirus (after SARS-CoV and MERS-CoV) and originated from bats. However, this novel coronavirus has been the only one having pandemic potential3, 4, 5, 6. SARS-CoV-2, a beta coronavirus, is an enveloped single-stranded, positive-sense, non-segmented and genetically diverse RNA virus with the largest genome size among known RNA viruses (29,891 ase pair, encodes for approximately 9860 amino acids)2, 7, 8. The genome of SARS-CoV-2 encodes both structural proteins like spike (S), envelope (E), membrane (M), and nucleocapsid (N), as well as non-structural proteins ranging from NSP1 to NSP16.
RNA viruses, generally, show a drastically high rate of mutation, substantially higher than those of DNA viruses. Due to this high rate of mutation shown by SARS-CoV-2 over a short period, it has been observed that viruses exhibit genomic variability which enables them to modulate virulence properties in the host and subsequently evade the host immunity9, 10.
In the present research work, we detected 987 mutations from 729 sequences derived from Asia in in the October. Altogether spike showed the highest mutations with 807 point mutations among the four structural proteins, followed by nucleocapsid with 151 mutations. Envelope showed 19 mutations and membrane showed only 10 point mutations. The results of our study suggest that mutational analysis of this virus might be considered as a new approach to help understand its genomic variability. Similarly, using the predictive tools of immunoinformatics approach, the antigenicity and allergenicity of the structural proteins of SARS-CoV-2 have been determined to develop efficacious antiviral therapeutics or vaccines against COVID-19.
Methods
Data mining
The full-length protein sequences of SARS-CoV-2 structural proteins, i.e., envelope protein, nucleocapsid phosphoprotein, surface glycoprotein and membrane glycoprotein, were retrieved from the NCBI virus database, as submitted from Asia in the month of October. There were 729 SARS-CoV-2 structural protein sequences submitted from Asia in the month of October, including sequences of 165 envelope proteins, 159 nucleocapsid phosphoproteins, 246 surface glycoproteins, and 159 membrane glycoproteins. A total of four reference sequences for envelope protein (YP_009724392), nucleocapsid phosphoprotein (YP_009724397), surface glycoprotein (YP_009724390), and membrane glycoprotein (YP_009724393) were also retrieved for mutational studies.
Multiple sequence alignment (MSA) and mutational identification
Multiple sequence alignment was performed using Clustal Omega online platform (http://www.clustal.org/) based on HMM profile seeded guide trees11. The envelope, nucleocapsid phosphoprotein, surface glycoprotein, and membrane glycoprotein were aligned with their respective reference sequences. The aligned files were viewed using Jalview (https://www.jalview.org/) to identify the point mutations occurring in different structural proteins with respect to the Wuhan type isolate.
Antigenicity and allergenicity evaluation
Vaxijen v2.0 server was used for the estimation of antigenicity of all the four structural proteins to study the capability of structural proteins to be used in vaccine production. This online server predicts antigens as per the auto cross-covariance (called ACC transformation) of the peptide sequences submitted to it12. A good vaccine needs to be non-allergenic to the host, hence the rationale for evaluating the allergenicity of these structural proteins, AllerTOP server was used, which predicts allergenicity based on size, flexibility, and other parameters13.
| Serial No. | Accession | Mutated sequence and position |
| 1. | BCM16104 | S68F |
| 2. | BCM16116 | S68F |
| 3. | BCM16128 | S68F |
| 4. | BCM16176 | S68F |
| 5. | BCM16188 | S68F |
| 6. | BCM16200 | S68F |
| 7. | BCM16212 | S68F |
| 8. | BCM16140 | S68F |
| 9. | QOP57282 | V75F |
| 10. | QOP57300 | V75F |
| 11. | QOP57289 | V75F |
| 12. | QOP57280 | V75F |
| 13. | QOP57294 | V75F |
| 14. | QOS50800 | V75F |
| 15. | QOS50895 | I46V |
| 16. | QOS50728 | V75F |
| 17. | QOS50501 | V75F |
| 18. | QOU99241 | I46V |
| 19. | QOU99253 | I46V |
| Serial No. | Accession | Mutated sequence and position |
| 1. | QJF74875 | R203K |
| 2. | QJF74875 | G204R |
| 3. | QKM75385 | R203K |
| 4. | QKM75385 | G204R |
| 5. | QKM75397 | R203K |
| 6. | QKM75397 | G204R |
| 7. | QKM75409 | R203K |
| 8. | QKM75409 | G204R |
| 9. | QKM75421 | R203K |
| 10. | QKM75421 | G204R |
| 11. | QKM75433 | R203K |
| 12. | QKM75433 | G204R |
| 13. | QKM75445 | P207L |
| 14. | QKM75445 | M210I |
| 15. | QKM75505 | R203K |
| 16. | QKM75505 | G204R |
| 17. | QKM75505 | D377G |
| 18. | QKM75517 | R203K |
| 19. | QKM75517 | G204R |
| 20. | QKM75517 | D377G |
| 21. | QKM75529 | R203K |
| 22. | QKM75529 | G204R |
| 23. | QKM75529 | D377G |
| 24. | QKM75541 | G204R |
| 25. | QKM75541 | D377G |
| 26. | QKM75541 | R203K |
| 27. | QKM75552 | R203K |
| 28. | QKM75552 | G204R |
| 29. | QKM75552 | D377G |
| 30. | QKM75563 | R203K |
| 31. | QKM75563 | G204R |
| 32. | QKM75563 | D377G |
| 33. | QKM75575 | R203K |
| 34. | QKM75575 | G204R |
| 35. | QKM75587 | R203K |
| 36. | QKM75587 | G204R |
| 37. | QKM75599 | R203K |
| 38. | QKM75599 | G204R |
| 39. | QKM75647 | R203K |
| 40. | QKM75647 | G204R |
| 41. | QKM75659 | R203K |
| 42. | QKM75659 | R204R |
| 43. | QKM75683 | R203K |
| 44. | QKM75683 | G204R |
| 45. | QKM75695 | R203K |
| 46. | QKM75695 | G204R |
| 47. | QKQ30536 | R203K |
| 48. | QKQ30536 | G204R |
| 49. | QKQ30548 | R40C |
| 50. | QKQ30560 | R203K |
| 51. | QKQ30560 | G204R |
| 52. | QKQ30572 | R203K |
| 53. | QKQ30572 | G204R |
| 54. | QKQ30584 | R203K |
| 55. | QKQ30584 | G204R |
| 56. | QLA10246 | R203K |
| 57. | QLA10246 | G204R |
| 58. | QLA10270 | R203K |
| 59. | QLA10270 | G204RR |
| 60. | QLA10282 | R203K |
| 61. | QLA10282 | G204R |
| 62. | QLA10294 | P383L |
| 63. | QLA10294 | R203K |
| 64. | QLA10294 | G204R |
| 65. | QLA10306 | R203K |
| 66. | QLA10306 | G204R |
| 67. | QLA10318 | G204R |
| 68. | QLA10318 | R203K |
| 69. | QLA10330 | G204R |
| 70. | QLA10330 | R203K |
| 71. | QLA10342 | G204R |
| 72. | QLA10342 | R203K |
| 73. | QLA10354 | R203K |
| 74. | QLA10354 | G204R |
| 75. | QOI53600 | P13L |
| 76. | QOQ57020 | S194L |
| 77. | QOQ57032 | S194L |
| 78. | QOQ57044 | S194L |
| 79. | QOQ57056 | S194L |
| 80. | QOQ57068 | S194L |
| 81. | QOQ57092 | M234I |
| 82. | QOQ57104 | S194L |
| 83. | QOQ57116 | S194L |
| 84. | QOQ57129 | S194L |
| 85. | QOQ72552 | S194L |
| 86. | QOQ72564 | S194L |
| 87. | QOQ72576 | S194L |
| 88. | QOQ84803 | S194L |
| 89. | QOQ84834 | S194L |
| 90. | QOR63442 | T205I |
| 91. | QOR63454 | S194L |
| 92. | QOR63466 | T205I |
| 93. | QOR63514 | A119S |
| 94. | QOR63514 | S194L |
| 95. | QOR64241 | S194L |
| 96. | QOR64253 | S194L |
| 97. | QOS50459 | P13L |
| 98. | QOS50495 | T91I |
| 99. | QOS50507 | P13L |
| 100. | QOS50519 | P13L |
| 101. | QOS50531 | P13L |
| 102. | QOS50590 | P13L |
| 103. | QOS50650 | P13L |
| 104. | QOS50674 | P13L |
| 105. | QOS50686 | P13L |
| 106. | QOS50686 | D225Y |
| 107. | QOS50722 | P13L |
| 108. | QOS50734 | P13L |
| 109. | QOS50746 | P13L |
| 110. | QOS50746 | S413I |
| 111. | QOS50758 | S413I |
| 112. | QOS50758 | P13L |
| 113. | QOS50770 | P13L |
| 114. | QOS50782 | P13L |
| 115. | QOS50818 | P13L |
| 116. | QOS50830 | P13L |
| 117. | QOS50853 | P13L |
| 118. | QOS50865 | P13L |
| 119. | QOS50889 | Q9H |
| 120. | QOS50889 | P199S |
| 121. | QOS50901 | S202N |
| 122. | QOS50924 | S202N |
| 123. | QOS50948 | P13L |
| 124. | QOS50960 | P13L |
| 125. | QOS50972 | P13L |
| 126. | QOS50996 | P13L |
| 127. | QOS51008 | P13L |
| 128. | QOS51020 | P13L |
| 129. | QOS51032 | P13L |
| 130. | QOS51068 | R209I |
| 131. | QOS51068 | P367L |
| 132. | QOS51080 | R203K |
| 133. | QOS51080 | G204R |
| 134. | QOS51092 | P13L |
| 135. | QOS51104 | R203K |
| 136. | QOS51104 | G204R |
| 137. | QOU99154 | P14L |
| 138. | QOU99201 | Q9H |
| 139. | QOU99201 | P199S |
| 140. | QOU99223 | Q9H |
| 141. | QOU99223 | P199S |
| 142. | QOU99247 | S202N |
| 143. | QOU99259 | S202N |
| 144. | QOU99270 | Q9H |
| 145. | QOU99270 | P199S |
| 146. | QOU99281 | Q9H |
| 147. | QOU99281 | P199S |
| 148. | QOU99292 | Q9H |
| 149. | QOU99292 | P199S |
| 150. | QOU99303 | Q9H |
| 151. | QOU99303 | P199S |
Results
Mutational identification
A total of 729 structural protein sequences were retrieved from the NCBI virus database for spike glycoproteins, nucleocapsid phosphoproteins, envelope proteins, and membrane glycoproteins submitted from Asian countries in the month of October 2020, along with four references sequences. The size of the different reference structural proteins, i.e., spikes glycoprotein, nucleocapsid phosphoprotein, envelope protein, and membrane glycoprotein being 1273, 419, 75, and 222 amino acids.
The sequences were viewed using Jalview after alignment to compare and detect the mutations among the Asian isolates with the Wuhan isolates with respect to structural proteins. Amongst the 729 sequences released from Asia, a total of 987 point mutations were detected in all four structural proteins (Figure 1). Among the 311 mutants, spike showed the highest mutations with 807 point mutations (Table 3), followed by nucleocapsid with 151 mutations (Table 2), while envelope showed 19 mutations (Table 1) and membrane showed only 10 point mutations (Table 4).
| S. No. | Accession | Mutated sequence and position |
| 1. | QJF74843 | V367F |
| 2. | QJF74867 | D614G |
| 3. | QOI53592 | M153I |
| 4. | QMI57728 | T95I |
| 5. | QMI57728 | N185K |
| 6. | QOI53580 | D614G |
| 7. | QOR64233 | D614G |
| 8. | QOR64245 | D614G |
| 9. | QOQ57012 | D614G |
| 10. | QOQ57024 | D614G |
| 11. | QOQ57060 | D614G |
| 12. | QOR64233 | A701T |
| 13. | QOR64233 | P812L |
| 14. | QOR64245 | P812L |
| 15. | QOQ57012 | P812L |
| 16. | QOQ57060 | P812L |
| 17. | QOR64233 | H1083Q |
| 18. | QOR64245 | H1083Q |
| 19. | QOQ57012 | H1083Q |
| 20. | QOQ57024 | H1083Q |
| 21. | QOQ57072 | D614G |
| 22. | QOQ57084 | D614G |
| 23. | QOQ57096 | D614G |
| 24. | QOQ57108 | D614G |
| 25. | QOQ57121 | D614G |
| 26. | QOQ57108 | A701T |
| 27. | QOQ57072 | A701T |
| 28. | QOQ57096 | P812L |
| 29. | QOQ57121 | P812L |
| 30. | QOQ57096 | H1083Q |
| 31. | QOQ57121 | H1083Q |
| 32. | QOQ57108 | H1083Q |
| 33. | QOQ72544 | L54F |
| 34. | QOQ72556 | L54F |
| 35. | QOQ72568 | L54F |
| 36. | QOQ72544 | D614G |
| 37. | QOQ72556 | D614G |
| 38. | QOQ72568 | D614G |
| 39. | QOQ72580 | D614G |
| 40. | QOQ84795 | D614G |
| 41. | QOQ72544 | A701T |
| 42. | QOQ72556 | P812L |
| 43. | QOQ72568 | P812L |
| 44. | QOQ72580 | P812L |
| 45. | QOQ72544 | H1083Q |
| 46. | QOQ84826 | D614G |
| 47. | QOR63434 | D614G |
| 48. | QOR63446 | D614G |
| 49. | QOR63458 | D614G |
| 50. | QOR63470 | D614G |
| 51. | QOR63434 | P812L |
| 52. | QOR63470 | P812L |
| 53. | QOQ53335 | S305T |
| 54. | QOQ53335 | C488R |
| 55. | QOR63482 | D614G |
| 56. | QOQ57036 | D614G |
| 57. | QOQ57048 | D614G |
| 58. | QOR63506 | D614G |
| 59. | QOQ53335 | D614G |
| 60. | QOQ57048 | A701T |
| 61. | QOQ57036 | P812L |
| 62. | QOQ57048 | P812L |
| 63. | QOQ57036 | H1083Q |
| 64. | QOQ57048 | H1083Q |
| 65. | QOQ53339 | F2L |
| 66. | QOQ53339 | V11I |
| 67. | QOQ53339 | S13R |
| 68. | QOQ53339 | Q14H |
| 69. | QOQ53339 | R34H |
| 70. | QOQ53339 | V42I |
| 71. | QOQ53339 | R44K |
| 72. | QOQ53339 | V47I |
| 73. | QOQ53339 | F59I |
| 74. | QOQ53339 | K77N |
| 75. | QOQ53339 | D111N |
| 76. | QOQ53339 | Q115H |
| 77. | QOQ53339 | A123T |
| 78. | QOQ53339 | N487I |
| 79. | QOQ53339 | V512L |
| 80. | QOQ53339 | A522P |
| 81. | QOQ53339 | A262T |
| 82. | QOQ53339 | Q677H |
| 83. | QOQ53336 | G199R |
| 84. | QOQ53340 | A262T |
| 85. | QOQ53338 | C301S |
| 86. | QOQ53340 | R328T |
| 87. | QOQ53337 | R457T |
| 88. | QOQ53338 | D614G |
| 89. | QOQ53340 | D614G |
| 90. | QOQ53336 | A684V |
| 91. | QOQ53336 | A688P |
| 92. | QOQ53336 | V705I |
| 93. | QOQ53337 | H1048Y |
| 94. | QOQ53337 | Q1180H |
| 95. | QOQ53337 | K1181Q |
| 96. | QOQ53341 | V11I |
| 97. | QOL24227 | V11I |
| 98. | QOQ53341 | R44K |
| 99. | QOQ53341 | V47I |
| 100. | QOL24227 | K77N |
| 101. | QOQ53341 | K77N |
| 102. | QOQ53341 | K97N |
| 103. | QOQ53341 | D111N |
| 104. | QOL24227 | D111N |
| 105. | QOL24227 | R190K |
| 106. | QOL24227 | D198E |
| 107. | QOL24225 | E224K |
| 108. | QOL24225 | D228N |
| 109. | QOL24226 | E224K |
| 110. | QOL24226 | D228N |
| 111. | QOL24227 | A262T |
| 112. | QOQ53341 | Q271H |
| 113. | QOQ53341 | F275L |
| 114. | QOL24228 | V407L |
| 115. | QOL24228 | P412S |
| 116. | QOL24227 | D427H |
| 117. | QOL24227 | N440H |
| 118. | QOL24227 | Q474P |
| 119. | QOL24228 | D614G |
| 120. | QOL24227 | D614G |
| 121. | QOQ53341 | G669R |
| 122. | QOQ53341 | Q675R |
| 123. | QOQ53341 | Q677H |
| 124. | QOL24227 | S686I |
| 125. | QOL24227 | A688P |
| 126. | QOL24225 | K790Q |
| 127. | QOL24226 | K790Q |
| 128. | QOL24225 | R815K |
| 129. | QOL24226 | R815K |
| 130. | QOL24225 | D820N |
| 131. | QOL24226 | D820N |
| 132. | QOL24225 | D830N |
| 133. | QOL24226 | D830N |
| 134. | QOL24228 | P863H |
| 135. | QOL24241 | F2L |
| 136. | QOL24241 | V11I |
| 137. | QOL24241 | Q14H |
| 138. | QOL24241 | R34C |
| 139. | QOL24241 | Y37N |
| 140. | QOL24241 | V42I |
| 141. | QOL24241 | R44K |
| 142. | QOL24241 | F65I |
| 143. | QOL78311 | S94F |
| 144. | QOL78311 | T95P |
| 145. | QOL24240 | D111N |
| 146. | QOL24240 | A282T |
| 147. | QOL78311 | D568H |
| 148. | QOL78311 | D614G |
| 149. | QOL24241 | H655Y |
| 150. | QOL24240 | Q675RR |
| 151. | QOL79057 | S13N |
| 152. | QOL79057 | D40E |
| 153. | QOL79057 | V42L |
| 154. | QOL79057 | S161F |
| 155. | QOL79057 | S246N |
| 156. | QOL79057 | D614G |
| 157. | QOL79058 | D614G |
| 158. | QOL79058 | R1019K |
| 159. | QOL79058 | P1090L |
| 160. | QOL79059 | V11F |
| 161. | QOL79059 | R21K |
| 162. | QOL79135 | R21K |
| 163. | QOL79135 | A222V |
| 164. | QOL79061 | K529I |
| 165. | QOL79059 | D614G |
| 166. | QOL79135 | D614G |
| 167. | QOL79061 | E619K |
| 168. | QOL79061 | G652R |
| 169. | QOL79061 | Q677H |
| 170. | QOL79061 | Y695N |
| 171. | QOL79061 | V729A |
| 172. | QOL79136 | D614G |
| 173. | QOL79137 | V11I |
| 174. | QOL79137 | Q115H |
| 175. | QOL79137 | D614G |
| 176. | QOL79137 | P863H |
| 177. | QOL79137 | Q913H |
| 178. | QOL79137 | I934T |
| 179. | QOL79333 | C136W |
| 180. | QOL79333 | N137Y |
| 181. | QOL79333 | I203L |
| 182. | QOL21485 | H207P |
| 183. | QOL20612 | E224K |
| 184. | QOL21486 | E224K |
| 185. | QOL21486 | R237K |
| 186. | QOL21486 | F238V |
| 187. | QOL21486 | Q239P |
| 188. | QOL20612 | T240S |
| 189. | QOL79332 | A262T |
| 190. | QOL79333 | L252P |
| 191. | QOL79332 | D467N |
| 192. | QOL21486 | A475V |
| 193. | QOL20612 | Q506H |
| 194. | QOL20612 | V510E |
| 195. | QOL20612 | V512E |
| 196. | QOL20612 | D614G |
| 197. | QOL21485 | V511I |
| 198. | QOL79333 | D568H |
| 199. | QOL79332 | Q675R |
| 200. | QOL20612 | V826G |
| 201. | QOL21485 | V826G |
| 202. | QOL20612 | I844M |
| 203. | QOL21486 | I844F |
| 204. | QOL21486 | R847K |
| 205. | QOL21486 | D848N |
| 206. | QOL21486 | C851W |
| 207. | QOL20612 | R847K |
| 208. | QOL20612 | D848N |
| 209. | QOL21535 | T21I |
| 210. | QOL21535 | V42A |
| 211. | QOL21535 | V511E |
| 212. | QOL21535 | C525W |
| 213. | QOL21535 | K557R |
| 214. | QOL21535 | R567K |
| 215. | QOL21535 | N657I |
| 216. | QOL21535 | Q677H |
| 217. | QOL21535 | V722A |
| 218. | QOL21535 | D737H |
| 219. | QOL21535 | T768P |
| 220. | QOL21535 | G769E |
| 221. | QOL21535 | E780K |
| 222. | QOL21535 | V781F |
| 223. | QOL21535 | K790Q |
| 224. | QOL21535 | I834J |
| 225. | QOL21536 | E224K |
| 226. | QOL21536 | V826G |
| 227. | QOL21536 | Y837N |
| 228. | QOL21536 | R847K |
| 229. | QOL21536 | D848N |
| 230. | QOL21536 | C851W |
| 231. | QOL21536 | L858F |
| 232. | QOL21681 | D228N |
| 233. | QOL21681 | E780K |
| 234. | QOL21681 | D808N |
| 235. | QOL21681 | S816P |
| 236. | QOL21681 | D820N |
| 237. | QOL21720 | T22I |
| 238. | QOL21720 | V213G |
| 239. | QOL21720 | E224K |
| 240. | QOL21720 | C538W |
| 241. | QOL21720 | K825Q |
| 242. | QOL10078 | R402I |
| 243. | QOL10078 | V407L |
| 244. | QOL10078 | P412S |
| 245. | QOL10078 | D614G |
| 246. | QOL10078 | P863H |
| 247. | QOJ86680 | T22I |
| 248. | QOK36756 | Y160I |
| 249. | QOK36756 | N185I |
| 250. | QOK36756 | Y200N |
| 251. | QOK36756 | D228Y |
| 252. | QOJ86685 | L226I |
| 253. | QOJ86685 | V227M |
| 254. | QOJ86685 | D228I |
| 255. | QOJ86685 | L249S |
| 256. | QOJ86685 | T250F |
| 257. | QOK36765 | E224K |
| 258. | QOK36765 | T240S |
| 259. | QOJ86685 | A263S |
| 260. | QOJ86684 | A262T |
| 261. | QOJ86684 | D287Y |
| 262. | QOJ86684 | V289E |
| 263. | QOK36765 | A522T |
| 264. | QOK36766 | V511I |
| 265. | QOJ86685 | P464L |
| 266. | QOJ86685 | T500I |
| 267. | QOJ86685 | N501R |
| 268. | QOJ86685 | R509K |
| 269. | QOJ86685 | V511E |
| 270. | QOJ86685 | S514P |
| 271. | QOJ86685 | H519I |
| 272. | QOJ86685 | P521Q |
| 273. | QOJ86685 | A522P |
| 274. | QOJ86685 | T523P |
| 275. | QOJ86685 | P527T |
| 276. | QOJ86685 | D568N |
| 277. | QOJ86685 | D627N |
| 278. | QOJ86684 | E583A |
| 279. | QOJ86684 | D586E |
| 280. | QOJ86684 | T602K |
| 281. | QOJ86684 | N603K |
| 282. | QOJ86684 | N606Y |
| 283. | QOJ86684 | C617R |
| 284. | QOJ86684 | G652R |
| 285. | QOJ86684 | E661K |
| 286. | QOJ86685 | E661D |
| 287. | QOJ86684 | A672P |
| 288. | QOK36766 | Q675R |
| 289. | QOK36766 | N824K |
| 290. | QOK36766 | K825Q |
| 291. | QOK36766 | V826M |
| 292. | QOK36765 | R847K |
| 293. | QOK36765 | D848N |
| 294. | QOK36765 | P863H |
| 295. | QOJ86680 | P863H |
| 296. | QOJ86680 | T874P |
| 297. | QOJ86680 | L877M |
| 298. | QOK36756 | P863H |
| 299. | QOK36767 | V11I |
| 300. | QOK36767 | R44K |
| 301. | QOK36767 | A684V |
| 302. | QOK36767 | V705I |
| 303. | QOK51484 | V11I |
| 304. | QOK51484 | K77N |
| 305. | QOK51484 | D111N |
| 306. | QOK51484 | Q115H |
| 307. | QOK51484 | V159L |
| 308. | QOK51484 | K187Q |
| 309. | QOK51484 | R190K |
| 310. | QOK51484 | K206N |
| 311. | QOK51484 | A262T |
| 312. | QOK51484 | D614G |
| 313. | QOK51484 | V620F |
| 314. | QOK51484 | G648R |
| 315. | QOK51484 | A706R |
| 316. | QOL00258 | V11I |
| 317. | QOL00258 | K77N |
| 318. | QOL00258 | D111N |
| 319. | QOL00258 | Q271H |
| 320. | QOL00258 | S443C |
| 321. | QOL00258 | G669R |
| 322. | QOL00258 | Q677H |
| 323. | QOL00258 | Q675R |
| 324. | QOL00259 | R44I |
| 325. | QOL00259 | K77N |
| 326. | QOL00259 | D111N |
| 327. | QOL00259 | R158L |
| 328. | QOL00259 | S162I |
| 329. | QOL00259 | E169Q |
| 330. | QOL00259 | Q173H |
| 331. | QOL00259 | Q183H |
| 332. | QOL00259 | D614G |
| 333. | QOL00259 | S686T |
| 334. | QOL00259 | E702Q |
| 335. | QOL00259 | V705I |
| 336. | QOL00259 | I714L |
| 337. | QOL00263 | V11I |
| 338. | QOL00263 | K77N |
| 339. | QOL00263 | Q115H |
| 340. | QOL00263 | H146P |
| 341. | QOL00263 | E1569D |
| 342. | QOL00263 | D614G |
| 343. | QOL00263 | S750T |
| 344. | QOL00264 | V11I |
| 345. | QOL00264 | V42I |
| 346. | QOL00264 | K77N |
| 347. | QOL00264 | D111N |
| 348. | QOL00264 | H207L |
| 349. | QOL00264 | A262T |
| 350. | QOL00264 | N487I |
| 351. | QOL00264 | V512L |
| 352. | QOL00264 | A522P |
| 353. | QOL00264 | G669R |
| 354. | QOL00264 | Q677H |
| 355. | QOL00264 | Q675R |
| 356. | QOL00468 | R44I |
| 357. | QOL00468 | K77N |
| 358. | QOL00468 | D111N |
| 359. | QOL00468 | R158L |
| 360. | QOL00468 | S162I |
| 361. | QOL00468 | E169Q |
| 362. | QOL00468 | Q173H |
| 363. | QOL00468 | V407L |
| 364. | QOL00468 | R408I |
| 365. | QOL00468 | A411T |
| 366. | QOL00468 | D614G |
| 367. | QOL00468 | D627N |
| 368. | QOL00468 | A653P |
| 369. | QOL00468 | N658H |
| 370. | QOL00468 | I664L |
| 371. | QOL00469 | V11I |
| 372. | QOL00469 | S13R |
| 373. | QOL00469 | T29Q |
| 374. | QOL00469 | R34H |
| 375. | QOL00469 | V42I |
| 376. | QOL00469 | R44K |
| 377. | QOL00469 | V47I |
| 378. | QOL00469 | F59I |
| 379. | QOL00469 | F65I |
| 380. | QOL00469 | K77N |
| 381. | QOL00469 | D111N |
| 382. | QOL00469 | S112T |
| 383. | QOL00469 | Q115H |
| 384. | QOL00469 | A123T |
| 385. | QOL00469 | A262T |
| 386. | QOL00469 | N487I |
| 387. | QOL00469 | V512L |
| 388. | QOL00469 | A522P |
| 389. | QOL00469 | E661D |
| 390. | QOL00469 | G669R |
| 391. | QOL00469 | A672P |
| 392. | QOL00469 | Q677H |
| 393. | QOL00501 | E224K |
| 394. | QOL00501 | I235V |
| 395. | QOL00501 | K814Q |
| 396. | QOL00501 | D830N |
| 397. | QOL00501 | P863H |
| 398. | QOL00502 | V11I |
| 399. | QOL00502 | V47I |
| 400. | QOL00502 | R78K |
| 401. | QOL00502 | K77N |
| 402. | QOL00502 | D111N |
| 403. | QOL00502 | Q115H |
| 404. | QOL00502 | E156D |
| 405. | QOL00502 | R158P |
| 406. | QOL00502 | G648R |
| 407. | QOL00502 | G652R |
| 408. | QOL00502 | S758T |
| 409. | QOL00502 | V785L |
| 410. | QOL00502 | K814N |
| 411. | QOL00502 | K854N |
| 412. | QOL00503 | P9Q |
| 413. | QOL00503 | V11I |
| 414. | QOL00503 | S13R |
| 415. | QOL00503 | R102T |
| 416. | QOL00503 | L18P |
| 417. | QOL00503 | Y37N |
| 418. | QOL00503 | N74H |
| 419. | QOL00503 | G103R |
| 420. | QOL00503 | K77N |
| 421. | QOL00503 | R78K |
| 422. | QOL00503 | K97N |
| 423. | QOL00503 | A262T |
| 424. | QOL00503 | A653P |
| 425. | QOL00503 | N658H |
| 426. | QOL00503 | I664L |
| 427. | QOL00503 | R408T |
| 428. | QOL00503 | G413R |
| 429. | QOL00503 | D427N |
| 430. | QOL00503 | D614G |
| 431. | QOL00503 | D627N |
| 432. | QOJ75919 | V11I |
| 433. | QOJ75919 | L176F |
| 434. | QOJ75919 | L179P |
| 435. | QOJ75919 | A262T |
| 436. | QOJ75919 | Q498Y |
| 437. | QOJ75919 | P499T |
| 438. | QOJ75919 | T716P |
| 439. | QOJ75919 | I726F |
| 440. | QOJ75920 | S438C |
| 441. | QOJ75920 | N544T |
| 442. | QOJ75920 | H655Y |
| 443. | QOJ75920 | V705F |
| 444. | QOJ75920 | Q836H |
| 445. | QOJ75921 | R21K |
| 446. | QOJ75921 | R44K |
| 447. | QOJ75921 | D111N |
| 448. | QOJ75921 | S112P |
| 449. | QOJ75921 | A262T |
| 450. | QOJ75921 | T302A |
| 451. | QOJ75921 | D614G |
| 452. | QOJ75922 | A262T |
| 453. | QOJ75922 | N536K |
| 454. | QOJ75922 | T791P |
| 455. | QOJ75922 | K811N |
| 456. | QOJ75922 | T827A |
| 457. | QOJ75923 | N460I |
| 458. | QOJ75923 | K462N |
| 459. | QOJ75923 | P463L |
| 460. | QOJ75923 | I468L |
| 461. | QOJ75923 | G838D |
| 462. | QOJ75923 | G842A |
| 463. | QOJ75923 | D848A |
| 464. | QOJ75924 | E132V |
| 465. | QOJ75924 | V159I |
| 466. | QOJ75924 | S162N |
| 467. | QOJ75924 | E191D |
| 468. | QOJ75924 | I197T |
| 469. | QOJ75924 | Q173F |
| 470. | QOJ75924 | M177I |
| 471. | QOJ75924 | C1043G |
| 472. | QOJ75924 | G485S |
| 473. | QOJ75924 | L513F |
| 474. | QOJ75924 | D614G |
| 475. | QOJ75924 | G838A |
| 476. | QOJ75924 | T998P |
| 477. | QOJ75924 | Q1011H |
| 478. | QOJ75924 | A1015P |
| 479. | QOJ75924 | D178Y |
| 480. | QOJ75924 | V193L |
| 481. | QOJ75924 | K187N |
| 482. | QOJ75924 | N188H |
| 483. | QOJ75924 | R190I |
| 484. | QOJ75924 | F194L |
| 485. | QOJ75925 | Q564H |
| 486. | QOJ75925 | F565C |
| 487. | QOJ75925 | Q1180P |
| 488. | QOJ75926 | T22I |
| 489. | QOJ75926 | P225R |
| 490. | QOJ75926 | I231L |
| 491. | QOJ75926 | R237S |
| 492. | QOJ75926 | G667A |
| 493. | QOJ75926 | A672P |
| 494. | QOJ75926 | Q677H |
| 495. | QOJ75926 | L242P |
| 496. | QOJ75926 | R246S |
| 497. | QOJ75926 | A688T |
| 498. | QOJ75927 | F65Y |
| 499. | QOJ75927 | V120D |
| 500. | QOJ75927 | V127D |
| 501. | QOJ75927 | F133L |
| 502. | QOJ75927 | F135L |
| 503. | QOJ75927 | P139T |
| 504. | QOJ75927 | C166S |
| 505. | QOJ75928 | L244F |
| 506. | QOJ75928 | L533F |
| 507. | QOJ75928 | D809K |
| 508. | QOJ75929 | V11I |
| 509. | QOJ75929 | T22I |
| 510. | QOJ75929 | A262T |
| 511. | QOJ75929 | N30I |
| 512. | QOJ75929 | H66L |
| 513. | QOJ75929 | N487I |
| 514. | QOJ75929 | N74H |
| 515. | QOJ75929 | N641K |
| 516. | QOJ75930 | D614G |
| 517. | QOJ75931 | V11I |
| 518. | QOJ75931 | H245Q |
| 519. | QOJ75931 | S247N |
| 520. | QOJ75931 | D614G |
| 521. | QOJ75931 | R685G |
| 522. | QOJ75932 | D614G |
| 523. | QOJ75932 | W1102C |
| 524. | QOJ75932 | N1108K |
| 525. | QOJ75932 | T1116N |
| 526. | QOJ75933 | D614G |
| 527. | QOJ75934 | D614G |
| 528. | QOJ75934 | T827I |
| 529. | QOJ75934 | L828F |
| 530. | QOJ75935 | V11I |
| 531. | QOJ75935 | S13R |
| 532. | QOJ75935 | Q14H |
| 533. | QOJ75935 | D614G |
| 534. | QOJ75935 | R682P |
| 535. | QOJ75935 | S686N |
| 536. | QOJ75935 | E702K |
| 537. | QOJ75935 | N710T |
| 538. | QOJ75935 | I714T |
| 539. | QOJ75936 | F194V |
| 540. | QOJ75936 | D614G |
| 541. | QOJ75936 | F759S |
| 542. | QOJ75936 | N764T |
| 543. | QOJ75936 | E702K |
| 544. | QOJ75936 | V705F |
| 545. | QOJ75936 | T739S |
| 546. | QOJ75936 | I742L |
| 547. | QOJ75936 | T747P |
| 548. | QOJ75936 | G757R |
| 549. | QOJ75937 | L560P |
| 550. | QOJ75937 | D614G |
| 551. | QOJ75938 | G648R |
| 552. | QOJ75938 | E661D |
| 553. | QOJ75938 | G669R |
| 554. | QOJ75938 | A672P |
| 555. | QOJ75938 | Q677H |
| 556. | QOJ75939 | D614G |
| 557. | QOJ75940 | N440H |
| 558. | QOJ75940 | F486L |
| 559. | QOJ75940 | D614G |
| 560. | QOJ75941 | A262P |
| 561. | QOJ75941 | Y266N |
| 562. | QOJ75941 | D614G |
| 563. | QOJ42687 | V3D |
| 564. | QOJ42687 | L10I |
| 565. | QOJ42687 | V11I |
| 566. | QOJ42687 | Q14H |
| 567. | QOJ42687 | N544T |
| 568. | QOJ42687 | E702K |
| 569. | QOJ42687 | V705J |
| 570. | QOJ42687 | W64C |
| 571. | QOJ42687 | T385S |
| 572. | QOJ42687 | E619K |
| 573. | QOJ42687 | S686T |
| 574. | QOJ42687 | A688P |
| 575. | QOJ42687 | S689N |
| 576. | QOJ42687 | Y695N |
| 577. | QOJ42687 | S721T |
| 578. | QOJ42687 | E725K |
| 579. | QOJ42687 | L727P |
| 580. | QOJ42687 | V729A |
| 581. | QOJ42687 | M731K |
| 582. | QOJ42687 | T22I |
| 583. | QOJ42687 | Y28N |
| 584. | QOJ42687 | T29P |
| 585. | QOJ42687 | K41Q |
| 586. | QOE90911 | Y38C |
| 587. | QOE90911 | F86S |
| 588. | QOE90911 | T95I |
| 589. | QOE90911 | E96G |
| 590. | QOE90911 | D290N |
| 591. | QOE90911 | L296F |
| 592. | QOE90911 | L303V |
| 593. | QOE90911 | S151G |
| 594. | QOE90911 | N185K |
| 595. | QOE90911 | P272S |
| 596. | QOE90911 | A288P |
| 597. | QOE90912 | A222P |
| 598. | QOE90912 | V227I |
| 599. | QOE90912 | K424N |
| 600. | QOE90912 | P863H |
| 601. | QOE90912 | K535S |
| 602. | QOE90912 | I1183F |
| 603. | QOE90912 | D1199Y |
| 604. | QOE90914 | D614G |
| 605. | QOE90914 | Y636F |
| 606. | QOE90914 | L650S |
| 607. | QOE90914 | P863H |
| 608. | QOE90915 | H656Y |
| 609. | QOE83920 | K77N |
| 610. | QOE83920 | A262T |
| 611. | QOE83920 | N542T |
| 612. | QOE83920 | D614G |
| 613. | QOE83920 | V620F |
| 614. | QOE83972 | P225T |
| 615. | QOE83972 | A262T |
| 616. | QOE83972 | N487I |
| 617. | QOE83972 | D614G |
| 618. | QOE83972 | V620I |
| 619. | QOE83972 | P863H |
| 620. | QOE83973 | Q115H |
| 621. | QOE83973 | E156D |
| 622. | QOE83973 | V171L |
| 623. | QOE83973 | K182N |
| 624. | QOE83973 | A262T |
| 625. | QOE83973 | D614G |
| 626. | QOE83973 | S758I |
| 627. | QOE83973 | P863H |
| 628. | QOE83973 | N925Y |
| 629. | QOE83973 | G932A |
| 630. | QOE83973 | T941P |
| 631. | QOE83974 | S13R |
| 632. | QOE83974 | L18P |
| 633. | QOE83974 | A262T |
| 634. | QOE83974 | P863H |
| 635. | QOE83974 | I931T |
| 636. | QOE83974 | G932A |
| 637. | QOE83974 | S943T |
| 638. | QOE83974 | Y37N |
| 639. | QOE83974 | I100T |
| 640. | QOE83974 | R408T |
| 641. | QOE83974 | D427N |
| 642. | QOE83974 | Q580H |
| 643. | QOE83974 | Y636F |
| 644. | QOE83974 | N641I |
| 645. | QOE83974 | G648R |
| 646. | QOE83975 | K535E |
| 647. | QOE83975 | H824N |
| 648. | QOE83975 | G842C |
| 649. | QOE83975 | V1177E |
| 650. | QOE84134 | I231L |
| 651. | QOE84134 | T240N |
| 652. | QOE84134 | N280D |
| 653. | QOE84134 | A288P |
| 654. | QOE84134 | T302A |
| 655. | QOE84134 | V736L |
| 656. | QOE84134 | N751H |
| 657. | QOE84134 | S816A |
| 658. | QOE84134 | D820N |
| 659. | QOE84134 | I788F |
| 660. | QOE84134 | K811N |
| 661. | QOE84134 | S813P |
| 662. | QOE84134 | K814Q |
| 663. | QOE84221 | F106L |
| 664. | QOE84221 | Q134P |
| 665. | QOE84221 | N148T |
| 666. | QOE84221 | A262T |
| 667. | QOE84221 | P863H |
| 668. | QOE84221 | V510E |
| 669. | QOE84221 | K535E |
| 670. | QOE84221 | N969H |
| 671. | QOE84221 | F486L |
| 672. | QOE84221 | N487I |
| 673. | QOE84221 | Q493P |
| 674. | QOE84221 | N501K |
| 675. | QOE84222 | A262T |
| 676. | QOE84222 | S459Y |
| 677. | QOE84222 | D614G |
| 678. | QOE84223 | P225S |
| 679. | QOE84223 | L229F |
| 680. | QOE84223 | I231L |
| 681. | QOE84223 | P863H |
| 682. | QOE84223 | K1181Q |
| 683. | QOE84223 | D820N |
| 684. | QOE84223 | L821R |
| 685. | QOE84223 | F823C |
| 686. | QOE84223 | R815K |
| 687. | QOE84223 | S816T |
| 688. | QOE84223 | I818F |
| 689. | QOE84223 | G798C |
| 690. | QOE84223 | K814M |
| 691. | QOE84223 | G838A |
| 692. | QOE84223 | G842R |
| 693. | QOE84223 | D843E |
| 694. | QOE84223 | V826L |
| 695. | QOE84223 | A829E |
| 696. | QOE84223 | D830H |
| 697. | QOE84223 | I834T |
| 698. | QOE84224 | D398N |
| 699. | QOE84224 | K535G |
| 700. | QOE84224 | E702K |
| 701. | QOE84224 | V705F |
| 702. | QOE84224 | E725K |
| 703. | QOE84224 | T736P |
| 704. | QOE84224 | D775N |
| 705. | QOE84224 | P863H |
| 706. | QOE84224 | Y837N |
| 707. | QOE84224 | A879P |
| 708. | QOE84224 | E918Q |
| 709. | QOU99158 | D614V |
| 710. | QOU99170 | D614V |
| 711. | QOU99296 | P1263L |
| 712. | QOS50441 | K77M |
| 713. | QOS50451 | A701S |
| 714. | QOS50594 | D614G |
| 715. | QOS50606 | D614G |
| 716. | QOS50630 | D614G |
| 717. | QOS50654 | D614G |
| 718. | QOS50654 | D936H |
| 719. | QOS50678 | D627P |
| 720. | QOS50726 | K77M |
| 721. | QOS50750 | I624T |
| 722. | QOS50834 | F135L |
| 723. | QOS50845 | M731I |
| 724. | QOS50952 | K77M |
| 725. | QOS50964 | K77M |
| 726. | QOS50928 | D614G |
| 727. | QOS50928 | K1045N |
| 728. | QOS50988 | K77M |
| 729. | QOS51000 | K77M |
| 730. | QOS51012 | K77M |
| 731. | QOS51024 | K77M |
| 732. | QOS51036 | K77M |
| 733. | QOS51024 | D88H |
| 734. | QOS50988 | D1084Y |
| 735. | QOS51000 | D1084Y |
| 736. | QOS51012 | D1084Y |
| 737. | QOS51024 | D1084Y |
| 738. | QOS51036 | D1084Y |
| 739. | QOS51060 | D614G |
| 740. | QOS51072 | D614G |
| 741. | QOS51096 | D614G |
| 742. | QOS51108 | D614G |
| 743. | QOS51120 | D614G |
| 744. | QKM75377 | D614G |
| 745. | QKM75389 | D614G |
| 746. | QKM75401 | D614G |
| 747. | QKM75413 | D614G |
| 748. | QKM75425 | D614G |
| 749. | QKM75377 | T547I |
| 750. | QKM75413 | V213L |
| 751. | QKM75473 | I569S |
| 752. | QKM75437 | D614G |
| 753. | QKM75449 | D614G |
| 754. | QKM75461 | D614G |
| 755. | QKM75473 | D614G |
| 756. | QKM75485 | D614G |
| 757. | QKM75485 | F797C |
| 758. | QKM75485 | G799S |
| 759. | QKM75497 | D614G |
| 760. | QKM75509 | D614G |
| 761. | QKM75521 | D614G |
| 762. | QKM75533 | D614G |
| 763. | QKM75545 | D614G |
| 764. | QKM75556 | D614G |
| 765. | QKM75567 | D614G |
| 766. | QKM75579 | D614G |
| 767. | QKM75591 | D614G |
| 768. | QKM75603 | D614G |
| 769. | QKM75579 | H146Q |
| 770. | QKM75591 | H146Q |
| 771. | QKM75556 | Y680F |
| 772. | QKM75567 | G1167V |
| 773. | QKM75615 | D614G |
| 774. | QKM75627 | D614G |
| 775. | QKM75639 | D614G |
| 776. | QKM75651 | D614G |
| 777. | QKM75663 | D614G |
| 778. | QKM75639 | N211Y |
| 779. | QKM75651 | N211Y |
| 780. | QKM75627 | G769V |
| 781. | QKM75675 | D614G |
| 782. | QKM75687 | D614G |
| 783. | QKQ30528 | D614G |
| 784. | QKQ30540 | D614G |
| 785. | QKQ30552 | D614G |
| 786. | QKQ30528 | G142V |
| 787. | QKM75687 | N211Y |
| 788. | QKQ30552 | N211Y |
| 789. | QKQ30552 | T236S |
| 790. | QKM75675 | S256L |
| 791. | QKM75675 | V1264M |
| 792. | QKQ30564 | D614G |
| 793. | QKQ30576 | D614G |
| 794. | QLA10238 | D614G |
| 795. | QLA10250 | D614G |
| 796. | QLA10262 | D614G |
| 797. | QKQ30576 | N211V |
| 798. | QLA10274 | D614G |
| 799. | QLA10286 | D614G |
| 800. | QLA10298 | D614G |
| 801. | QLA10310 | D614G |
| 802. | QLA10322 | D614G |
| 803. | QLA10298 | G75V |
| 804. | QLA10322 | V1068F |
| 805. | QLA10334 | D614G |
| 806. | QLA10346 | D614G |
| 807. | QLA10346 | H146Q |
| Sl. No. | Accession | Mutated sequence and position |
| 1. | QOR64248 | S214R |
| 2. | QOQ57027 | S214R |
| 3. | QOQ57075 | S214R |
| 4. | QOQ72547 | S214R |
| 5. | QOQ84829 | S214R |
| 6. | QOQ57051 | S214R |
| 7. | QKM75606 | A2V |
| 8. | QKM75618 | A2V |
| 9. | QKQ30543 | A2V |
| 10. | QLA10253 | A2V |
| Structural protein | Antigenicity | Allergenicity |
| Surface glycoprotein | 0.4696 | Non-allergen |
| Envelope protein | 0.6025 | Non-allergen |
| Nucleocapsid phosphoprotein | 0.5059 | Non-allergen |
| Membrane glycoprotein | 0.5102 | Non-allergen |
Assessment of antigenicity and allergenicity
VaxiJen v2.0 server was used to predict the antigenicity of all four structural proteins of SARS-CoV-2. A peptide to be used in vaccine production must bind with the B-cell and T-cell receptors and enhance the cell's immune response. This estimation indicated that envelope protein was the most antigenic one among the four with an antigenicity of 0.6025, followed by membrane glycoprotein, having antigenicity of 0.5102. In contrast, the antigenicity of nucleocapsid phosphoprotein ranked third with a value of 0.5059, and surface glycoprotein was the least antigenic with antigenicity of 0.4696 as shown in Table 5. The allergenicity of these proteins was also estimated to discern whether these antigenic peptides were allergenic or not. A good vaccine should be non-allergenic to the host, and it should not produce any IgE-mediated immune responses in the host. The allergenicity analysis revealed that all four structural proteins were non-allergenic to the host and could be used as potent vaccine targets.
Discussion
On 11th March 2020, the WHO announced COVID-19 as a global pandemic due to its rapid global spread to numerous countries, and declared it as a serious threat to public health across the world14. Therefore, antiviral therapeutics or candidate vaccines were imminently necessary to curb SARS-CoV-2 infections. The virus primarily caused acute to severe respiratory illness and pneumonia in humans. The symptoms of COVID-19 began within two days, and could continue up to 14 days.
The emergence of a huge number of novel mutations in the genome of SARS-CoV-2 may interrupt ongoing vaccine development and trial strategies in different parts of the world. Regular monitoring of mutations has been crucial in tracking and tracing the circulation of this virus among individuals and in different geographical locations. SARS-CoV-2 (Wuhan-Hu-1), complete genome sequence, was deposited in the NCBI Gene bank15 in January 2020.
RNA viruses, including SARS-CoV-2, exhibit a high frequency of novel point mutations which are supposedly beneficial for the viruses to adapt and evolve in changing climatic conditions, thereby enhancing their potential transmission worldwide16, 17. The mutation is considered as the essential natural selection phenomenon and most often select for those traits of the virus that are a pre-requisite for survival in the highly dynamic host environment. These characteristic features of viruses (e.g. SARS-CoV-2) could complicate the ongoing efforts of researchers to combat this contagious disease because the high frequency of viral mutations induces drug resistance and rapid immune evasion18, 19.
The results of our study revealed the presence of a total of 987 mutations from 729 sequences from Asia countries in October. Spike glycoproteins were found to be highly mutagenic amongst the structural proteins, followed by nucleocapsid (151 mutations). Meanwhile, envelope protein showed 19 mutations and only 10 point mutations were found in the membrane proteins. Furthermore, we estimated the antigenicity and allergenicity of these structural proteins to design and develop potentially potent candidate vaccines against SARS-CoV-2. Among the structural proteins, envelope (E) was found to be highly antigenic and least allergenic, followed by nucleocapsid (N), membrane (M), and spike (S) protein of SARS-CoV-2. Antigenicity and allergenicity are the essential criteria to develop efficacious antiviral therapeutics or vaccines against COVID-19.
The mutations cause alterations in the structural proteins of the SARS-CoV-2 virus and, therefore, help in evading the host immunity. The occurrence of the high frequency of mutations creates a barrier in the development of antiviral drugs. Our results shown in this article, here presents a snap shot picture of an enormously changing situation due to SARS-CoV-2.
Conclusions
In the present study, the occurrence of novel mutations in the different structural proteins of the SARS-CoV-2 virus provides further insight into the identification and magnitude of virulence properties of virus strains, from a large repertoire of strains. Our findings might be useful in the development of effective therapeutic strategies against all types of SARS-CoV-2 strains.
Abbreviations
COVID-19: coronavirus disease 2019
E: envelope
M: membrane
N: nucleocapsid
S: spike
SARS: Severe acute respiratory syndrome
Acknowledgments
Not applicable
Author’s contributions
All authors read and approved the final manuscript.
Funding
None
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
References
-
Lu
R.,
Zhao
X.,
Li
J.,
Niu
P.,
Yang
B.,
Wu
H.,
Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet.
2020;
395
(10224)
:
565-74
.
View Article PubMed Google Scholar -
Zhu
N.,
Zhang
D.,
Wang
W.,
Li
X.,
Yang
B.,
Song
J.,
China Novel Coronavirus Investigating
Research Team
A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med.
2020;
382
(8)
:
727-33
.
View Article PubMed Google Scholar -
Zhang
T.,
Wu
Q.,
Zhang
Z.,
Pangolin homology associated with 2019-nCoV. BioRxiv.
2020
.
View Article Google Scholar -
Lau
S.K.,
Woo
P.C.,
Li
K.S.,
Huang
Y.,
Tsoi
H.W.,
Wong
B.H.,
Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA.
2005;
102
(39)
:
14040-5
.
View Article PubMed Google Scholar -
Alagaili
A.N.,
Briese
T.,
Mishra
N.,
Kapoor
V.,
Sameroff
S.C.,
Burbelo
P.D.,
Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio.
2014;
5
(2)
:
e00884-14
.
View Article PubMed Google Scholar -
Guan
Y.,
Zheng
B.J.,
He
Y.Q.,
Liu
X.L.,
Zhuang
Z.X.,
Cheung
C.L.,
Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science.
2003;
302
(5643)
:
276-8
.
View Article PubMed Google Scholar -
Cascella
M.,
Rajnik
M.,
Aleem
A.,
Dulebohn
S. C.,
Napoli
R. D.,
Features, Evaluation, and Treatment of Coronavirus (COVID-19). https://www.ncbi.nlm.nih.gov/books/NBK554776/.
2020
.
-
Barnard
D.L.,
Coronaviruses: molecular and cellular biology. Future Virol.
2008;
3
(2)
:
119-23
.
View Article PubMed Google Scholar -
N.S. Ogando,
F. Ferron,
E. Decroly,
B. Canard,
C.C. Posthuma,
E.J. Snijder,
The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity. Front Microbiol.
2019;
10
:
1813
.
View Article PubMed Google Scholar -
Eckerle
L.D.,
Becker
M.M.,
Halpin
R.A.,
Li
K.,
Venter
E.,
Lu
X.,
Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog.
2010;
6
(5)
:
e1000896
.
View Article PubMed Google Scholar -
Madeira
F.,
Park
Y.M.,
Lee
J.,
Buso
N.,
Gur
T.,
Madhusoodanan
N.,
The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res.
2019;
47
:
636-41
.
View Article PubMed Google Scholar -
Doytchinova
I.A.,
Flower
D.R.,
VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics.
2007;
8
(1)
:
4
.
View Article PubMed Google Scholar -
Dimitrov
I.,
Flower
D.R.,
Doytchinova
I.,
AllerTOP\textemdasha server for in silico prediction of allergens. BMC Bioinformatics.
2013;
14
:
4
.
View Article PubMed Google Scholar -
World Health Organization
WHO Bull 2020.
Google Scholar -
Wu
F.,
Zhao
S.,
Yu
B.,
Chen
Y.M.,
Wang
W.,
Song
Z.G.,
A new coronavirus associated with human respiratory disease in China. Nature.
2020;
579
(7798)
:
265-9
.
View Article PubMed Google Scholar -
Daniele
M.,
Federico
M.G.,
Front Microbio 2020.
Google Scholar -
Domingo
E.,
Holland
J.J.,
RNA virus mutations and fitness for survival. Annu Rev Microbiol.
1997;
51
(1)
:
151-78
.
View Article PubMed Google Scholar -
Irwin
K.K.,
Renzette
N.,
Kowalik
T.F.,
Jensen
J.D.,
Antiviral drug resistance as an adaptive process. Virus Evolution.
2016;
2
(1)
:
vew014
.
View Article Google Scholar -
Yashvardhini
N.,
Kumar
A.,
Jha
D.K.,
Immunoinformatics Identification of B- and T-Cell Epitopes in the RNA-Dependent RNA Polymerase of SARS-CoV-2. Canadian J Infec Diseases Med Microbiol.
2021;
2021
:
6627141
.
View Article Google Scholar
Comments
Downloads
Article Details
Volume & Issue : Vol 8 No 5 (2021)
Page No.: 4367-4381
Published on: 2021-05-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 5995 times
- Download downloaded - 1384 times
- View Article downloaded - 0 times
 Biomedpress
Biomedpress



