Abstract
Introduction: Hepatitis C virus (HCV) infection frequently leads to liver complications, such as fibrosis, cirrhosis, and hepatocellular carcinoma. The incidence of HCV infection transmission among drug abusers is concerning. Interleukin-1 beta (IL-1b) is a pro-inflammatory cytokine secreted during innate and adaptive immune responses and plays a pivotal role in chronic inflammatory diseases. Functional single nucleotide polymorphisms in IL-1b cause it to play different roles in disease susceptibility and progression. This study aimed to investigate the association between genetic polymorphisms of pro-inflammatory cytokines (IL-1b) and HCV infection susceptibility in Malay male drug abusers.
Methods: In total, 48 male Malay drug abusers were included in this retrospective case-control study. Genomic DNA was extracted from whole blood samples and analyzed using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) for the IL-1b rs16944 and rs1143634 polymorphisms.
Results: Analysis of IL-1b rs1143634 revealed that the C/C genotype was common in both the case and control groups; however, no statistical significance was observed (p = 0.068, c2 = 3.755). Genotyping of IL-1b demonstrated that all samples were of the homozygous mutant type (T/T).
Conclusion: There was no association between IL-1b polymorphism (rs1143634 and rs16944) and hepatitis C infection susceptibility among Malay male drug abusers.
INTRODUCTION
Hepatitis C virus (HCV) infection has been recognized as a major global public health issue since 2015. It is estimated that 71 million people have been diagnosed with HCV; currently, there are 58 million chronically infected individuals1. Although the decrease in the number of chronic infections is cause for optimism, it should not detract from the severity of this infection. According to the World Health Organization (2022), approximately 290,000 HCV-related deaths were in 2019. These deaths were primarily due to chronic infections progressing to cirrhosis and hepatocellular carcinoma (HCC), as 20% of persistent HCV infection cases develop these conditions2, 3. Up to 80% of acute HCV infections are asymptomatic, making it difficult to diagnose, which, in turn, increases the risk of latent infection transmission and progression to chronic infection4, 5.
HCV is a bloodborne liver disease. Injecting hazardous parenteral drugs, engaging in high-risk sexual activity, and receiving blood transfusions are the three most common modes of HCV transmission3, 6, 7, 8, 9. Therefore, healthcare patients, healthcare workers, and drug abusers are at the highest risk of contracting HCV. Due to this, drug abusers have been the focus of recent studies10. Hazardous activities, such as illicit drug use and the sharing of contaminated syringes, escalate HCV transmission11, 12.
HCV infection is endemic globally; however, the prevalence of the disease varies globally6, 13. Studies on the global distribution and prevalence of HCV genotypes revealed the diverse prevalence of genotypes 1, 2, 3, and 6 across Southeast Asian countries13, 14. Southeast Asia has a particularly high prevalence of genotype 6, with 94.6 million infected individuals15. Studies on HCV genotype prevalence in Malaysia reported that genotype 3 is the most common, followed by genotype 116, 17. In 2017, HCV infection had an incidence of 9.54 in 100,000 people, while the mortality rate was 0.29 per 100,000 people18.
Viral proteins and double-stranded RNA (dsRNA) from HCV induce the production of pro-inflammatory cytokines19. During the early stages of infection, the intricate cytokine network enables the formation of coordinated and efficient innate and adaptive immune responses20, 21, 22, 23. A viral infection outcome is determined by the interaction between the host’s capacity to generate a powerful antiviral response and the viral mechanisms that neutralize such responses24. It is impossible to adequately emphasize the significance of pro-inflammatory cytokines, such as interleukin-1 (IL-1), in immune response regulation. This is because many factors influence immune system function, particularly when it is affected by diseases such as HCV. Addressing the genetic diversity of a host during a latent infection is one way to ascertain the significant associations between the behaviors of different cytokines.
Functional single nucleotide polymorphisms (SNPs) of IL-1 have diverse distributions among major populations in America, Europe, South Asia, East Asia, and Africa. Studies have reported that different IL-1 expression levels and polymorphisms may impart different effects on disease conditions25, 26, 27, 28. Therefore, there is a growing need to understand the role that SNPs play in the progression of chronic HCV infection due to the growing body of evidence suggesting a relationship between SNPs and disease progression, particularly in diseases involving inflammation. Genotyping and SNP analysis provide information that can be used to build genome-based knowledge regarding individual susceptibility to a variety of common diseases, produce safer and more effective personalized diets and treatments for patients, and improve the understanding of the evolutionary processes of diseases29. This study aimed to investigate the association between the genetic polymorphisms rs16944 and rs1143634 of pro-inflammatory cytokine IL-1 and HCV infection susceptibility in male Malay drug abusers.
MATERIALS AND METHODS
Subject recruitment and DNA extraction
A retrospective case-control study design was used involving male Malay drug abusers. The sample size was calculated using a web calculator and in reference to a previous study30, 31. The sample size consisted of 29 male Malay drug abusers with chronic HCV infection (case group) and 19 male Malay drug abusers without HCV (control group). The participants’ ages ranged from 35–60 years old. Medical histories were obtained, and physical examinations were performed for each participant. Subject recruitment received ethical review and approval from the Medical Research and Ethics Committee, Ministry of Health Malaysia (NMRR-19-399-45866), and the USM Human Research Ethics Committee (USM/JEPeM/18010012).
Several inclusion criteria were used: 1) participants had to be third-generation Malay, 2) be at least 18 years old and have a history of drug abuse, and 3) for the control group, have tested positive for HCV for six months based on a structured clinical review using the DSM-V. Participants who met any of the exclusion criteria were not included in the analysis: 1) had non-HCV-related liver disease, 2) had a mental disorder, and 3) refused or were unable to provide informed consent.
A 10 mL blood sample was collected from each patient in a sterile tube containing heparin K2EDTA. Genomic DNA was extracted using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Spectrophotometry was used to determine the purity and concentration of the extracted DNA. DNA quality was reflected by a consistent ratio of 1.8 – 2.0. The coded genomic DNA solution was stored at 4°C.
Genotyping with PCR-RFLP
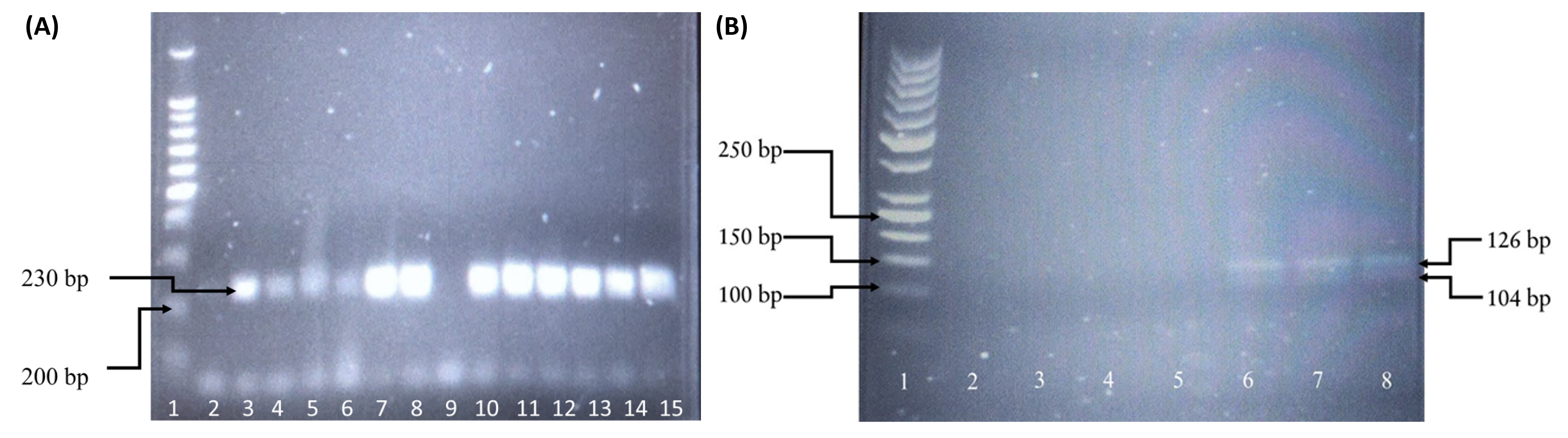
The extracted genomic DNA extracted was genotyped using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The amplification was performed in 25 μL of reaction solution containing a mixture of PCR buffer (1×), dNTPs (2 mM), MgCl2 (1.5 mM), forward and reverse primers (20 pmol) (Table 1), Taq polymerase (2.5 U), and sufficient nuclease-free water. PCR amplification targeting IL-1β rs16944 was performed following several steps: an initial denaturation at 95°C for 5 minutes followed by 35 cycles at 95°C for 30 seconds, 57°C for 30 seconds, 72°C for 30 seconds, and a final extension at 95°C for 5 minutes. The PCR product was then mixed with 2 µL of AvaI (5 U) and incubated at 37°C for 2 hours. Genotypes of rs16944 were identified using their base pair size, where the wildtype (C/C) corresponded to two bands of 190 and 116 base pairs, heterozygous (C/T) corresponded to three bands of 190, 116, and 306 base pairs, and mutant (T/T) corresponded to a single band of 306 base pairs. For rs1143634, different PCR steps were followed: an initial denaturation at 94°C for 5 minutes followed by 35 cycles at 94°C for 30 seconds, 58°C for 30 seconds, 72°C for 30 seconds, and a final extension at 94°C for 5 minutes. The PCR product was then mixed with 2 µL of TaqI (5 U) and incubated at 65°C for 2 hours. Genotypes were identified according to their base pair size, where wildtype (C/C) corresponded to two bands of 126 and 104 base pairs, heterozygous (C/T) corresponded to three bands of 126, 104, and 230 base pairs, and mutant (T/T) corresponded to a single band of 230 base pairs. Digestion fragments were separated by electrophoresis on 2% agarose with gel stain (Invitrogen SYBR Safe DNA, USA) using the available size ladder for comparison.
| RE | Forward Primers (5'-3') | Reverse Primers (5'-3') | Ref. | |
|---|---|---|---|---|
| rs16944 | AvaI | TGGCATTGATCTGGTTCATC | GTTTAGGAATCTTCCCACTT | 32 |
| rs1143634 | TaqI | GACTTTGACCGTATATGCTCAG | ATGGACCAGACATCACCAAGC | 33 |
Statistical analysis
The independent samples t-test and Mann–Whitney test were used to compare continuous data. The Hardy–Weinberg equilibrium (HWE) was used to ascertain the quality of the genotyping method for genotype and allelic analysis. The HWE p-value was determined by comparing the control group with a reference population in a 2×2 contingency table (CC vs. CT+TT). Differences in allele and genotype frequencies between the study groups were compared using chi‐squared tests. Statistical significance was defined as p < 0.05. Fischer’s exact test was employed in place of the chi-squared test when more than 20% of the expected counts were less than five in the 2×2 contingency table.
| Characteristics | Drug abuser | P-value | |
|---|---|---|---|
| Case Group (n = 29) | Control Group (n = 19) | ||
| Age (year) | 44.59 (6.23) | 43.05 (7.48) | 0.445a |
| Weight (kg) | 164.76 (4.76) | 164.37 (7.85) | 0.847a |
| Height (m) | 58.18 (7.33) | 59.58 (9.08) | 0.560a |
| BMI (kg/m 2 ) | 21.46 (2.67) | 22.02 (2.86) | 0.491a |
| Systolic BP (mm/Hg) | 120.55 (16.94) | 120.68 (10.68) | 0.378b |
| Diastolic BP (mm/Hg) | 82.37 (8.87) | 79.26 (7.73) | 0.221b |
| Bilirubin (µmol/L) | 9.89 (9.84) | 6.45 (2.8) | 0.176b |
| ALT (U/L) | 33.43 (18.91) | 42.31 (38.8) | 0.987b |
| AST (U/L) | 49.48 (24.64) | 48.15 (34.31) | 0.429b |
| ALP (U/L) | 103.39 (29.13) | 96.23 (31.61) | 0.497a |
| GGT (U/L) | 65.35 (42.03) | 79.85 (75.63) | 0.908b |
| Characteristics | Drug abuser | P value | |
|---|---|---|---|
| Case Group (n=29) | Control Group (n=19) N (%) | ||
| HIV co-infection | 17 (58.62) | 4 (21.05) | 0.01c* |
| IL-1β | Genotype and allele | Control group (n=19), N (%) | Case group (n=29), N (%) | EAS HWE (n=504), N (%) | HWE P-valued (<0.05) | χ2 (df) | p-value |
|---|---|---|---|---|---|---|---|
| -511 (C>T) rs16944 | CC | 0 | 0 | 145 (28.80) | 0.006* | - | - |
| CT | 0 | 0 | 245 (48.61) | ||||
| TT | 19 (100) | 29 | 114 (22.62) | 0.000* | - | - | |
| C | 0 | 0 | 535 (53.10) | ||||
| T | 38 (100) | 58 (100) | 473 (46.92) | ||||
| +3954 (C>T) rs1143634 | CC | 18 (94.74) | 21 (72.41) | 482 (95.63) | 0.581 | 3.755 (1) | 0.068 |
| CT | 1 (5.26) | 7 (24.14) | 21 (4.17) | ||||
| TT | 0 | 1 (3.45) | 1 (0.20) | ||||
| C | 37 (97.37) | 49 (84.48) | 985 (97.72) | 0.593 | 4.085 (1) | 0.083 | |
| T | 1 (2.63) | 9 (15.52) | 23 (2.28) |
RESULTS
Table 2 shows the demographic and clinical characteristics of the study participants. Table 3 shows the association between HCV infection and human immunodeficiency virus (HIV) co-infection in male Malay drug abusers. Figure 1 and Figure 2 present the genotyped samples targeting IL-1β at positions −511 (rs16944) and +3954 (rs1143634). The genotype and allele distributions of rs16944 and rs1143634 between the case and control groups are presented in Table 4.
No significant differences were seen in age, weight, height, body mass index (BMI), systolic blood pressure, and diastolic blood pressure between the case and control groups. In addition, liver test results (bilirubin level, AST, ALP, ALT, and GGT) did not significantly differ between the case and control groups. However, there was a significant correlation between HCV infection and HIV co-infection; 58.62% of case group participants were co-infected with HIV, while the control subjects had a co-infection rate of 21.05% (p = 0.01, χ2 = 6.583).
For rs16944, the HWE value deviated from the reference population (p < 0.05). Allelic analysis revealed similar findings. Genotyping of IL-1β rs16944 showed that all samples from the case and control groups were of the homozygous mutant type (T/T). Allelic frequency analysis indicated that the T allele was dominant. Due to the non-varied frequencies of alleles and genotypes, no analysis could be performed. The chi-squared test results and p-values of the genotypic and allelic frequency for this gene were undefined.
The HWE result for the genotyping of rs1143634 was reliable (p = 0.581). The frequency of this genotype in the control group was comparable to the reference population. Allelic analysis demonstrated similar results (p = 0.593). Genotyping of IL-1β rs1143634 revealed that 21 (72.41%) HCV-infected patients had the homozygous wildtype (C/C), 7 (24.14%) were heterozygous (C/T), and 1 (3.45%) was mutant (T/T). The frequencies between the reference population and the control group demonstrated similar distributions. The genotypic frequency of the homozygous wildtype was much higher in the control group than in the case group; the observed frequency of the C/C genotype in the control group was 94.74%. However, the heterozygous genotype in the control group was much lower than in the case group, with only one (5.26%) group participant demonstrating this genotype. There were no homozygous mutant (T/T) genotypes reported in the control group. The frequency of the C allele in the control group (37; 97.37%) was higher than in the case group (49; 84.48%), while the frequency of the T allele was higher in the case group (9; 15.52%) compared to the control group (1; 2.63%). Statistical evaluation of IL-1β rs1143634 revealed no significant association between HCV infection and genotype distribution (p = 0.068, χ2 = 3.755). Furthermore, there was no significant association between the allelic distribution and HCV infection (p = 0.083).
DISCUSSION
Since its discovery, HCV infection has been a continuous public health concern. The acute stage of the infection is typically followed by the progression to chronic disease if the infection is allowed to persist. There are several factors that affect HCV progression and an individual’s susceptibility to the transformation of an acute HCV infection into a chronic condition. For instance, the illicit use of controlled substances disrupts liver metabolism. This disruption occurs proportionally to the role of specific substances as immunological mediators. Drugs are well-known immune modulators, and excessive use of these substances can lead to elevated levels of inflammatory cytokines and an increased risk of infection34.
In the general population, the rs16944 SNP was found to exhibit a heterogenous genotype (C/T). The frequency of the T allele was found to be slightly higher than the C allele. In the East Asian (EAS) population, the most common genotype was the heterogenous (C/T) genotype. Furthermore, the C allele was slightly more prevalent in the EAS population. Comparing the allelic distribution in the control group and the EAS population, a significant deviation in genotypic and allelic distribution was found. Regarding the rs1143634 SNP, the C/C genotype was predominant in the general population. Additionally, the C allele frequency was increased in the general population. The C/C genotype was the most common within the EAS population, and the C allele had a higher frequency than the A allele. The rs1143634 genotype in the control group exhibited a similar trend to the reference population; the HWE values were in equilibrium and, therefore, did not deviate from the reference population.
In the absence of disrupting events, the HWE states that genetic variation in a population will remain constant from generation to generation. This principle is based on several assumptions, including random mating, the absence of natural selection, a large population size, no gene flow or migration, no mutation, and the autosomal nature of the locus35. The deviation from the HWE observed in the control group may be indicative of significant issues, such as selection bias, population stratification, and genotyping errors36, 37.
The results of the current study indicated no relationship between the genotypic frequency of IL-1β rs1143634 and HCV infection. Furthermore, the frequency of homozygous C/C—which corresponds to the G/G genotype—was higher in the control group. The frequency of the C allele was not significantly correlated with chronic HCV illness. In a previous study, no genotypes of IL-1β rs1143634 demonstrated a significant association with the progression to chronic HCV infection among Tunisian patients38. However, the C/C genotype frequency in the G2 group (62.5%) was higher than the frequency of heterozygous C/T and homozygous mutant (T/T) genotypes. The G2 group was comprised of 24 participants who were thought to have recovered from HCV infection without medical intervention, as evidenced by two HCV-PCR tests taken one year apart that were both negative39.
Previous studies have found that IL-1β rs1143634 genotypes have no significant influence on liver cirrhosis predisposition in patients with chronic HCV infection40. The genotypic distribution of IL-1β rs1143634 has also been shown to have no influence on spontaneous HCV clearance among the European population41. Another study on the genotypic frequencies of the IL-1β rs1143634 gene and HCV treatment inferred that the C/C genotype was strongly associated with interferon therapy38. In addition, the examination of allelic distribution revealed that the T allele was associated with treatment non-response, increased fibrosis, and increased hepatic activity. In contrast, the C allele was significantly associated with decreased fibrosis, lower hepatic activity, and persistent viral response.
According to existing research39, 42, 43, 44, 45, 32, 33, polymorphisms of IL-1β at locus −511 are associated with progression to chronic HCV infection. However, the genotypes associated with HCV infection susceptibility are diverse. While one study found that the C/C genotype was predominant in HCV-positive patients42, another reported that the C/T genotype was more prevalent in Wang et al.'s study (2003)43. Moreover, other research has concluded that the T/T genotype was a significant risk factor for the progression of chronic HCV infection to HCC44. In addition, the mutant T/T allele has been frequently reported in patients with chronic HCV and HCV-induced liver cirrhosis42. An extensive inflammasome gene polymorphism study reported that the C allele of IL-1β rs16944 was less likely to advance hepatitis C development45. These differences in IL-1β rs16944 genotypes are primarily caused by the ethnic background of the population observed in each respective study, namely, American, Asian, and European
In Malaysia, there is currently a lack of research regarding the effects of IL-1β gene polymorphisms (IL-1β rs16944 and rs1143634) in drug abusers with chronic HCV infection.
CONCLUSIONS
There was no association between IL-1β polymorphisms (rs1143634 and rs16944) and HCV infection susceptibility among Malay male drug abusers. The partial results of IL-1β genotype and allele frequency analysis are in line with the findings of several previous studies; however, some previous studies contradict the current findings.
Abbreviations
ALP: Alkaline phosphatase, ALT: Alanine aminotransferase, AST: Aspartate transaminase, BMI: Body Mass Index, DNA: Deoxyribonucleic acid, DSM-V: Diagnostic and Statistical Manual of Mental Disorders, dsRNA: Double strand ribonucleic acid, EAS: East Asian, GGT: Gamma-glutamyl transpeptidase, HCC: Hepatocellular carcinoma, HCV: Hepatitis C virus, HIV: Human Immunodeficiency Virus, HWE: Hardy-Weinberg Equilibrium, IL-1β: Interleukin 1- beta, PCR-RFLP: Polymerase Chain Reaction Restriction Fragment Length Polymorphism, RNA: Ribonucleic acid, SNP: Single nucleotide polymorphism, WHO: World Health Organization
Acknowledgments
We would like to express our gratitude to the Kelantan State Health Department and Kota Bharu District Health Office for their cooperation. We also thank the staffs at Health Clinics in Kota Bharu District for their assistance in this study.
Author’s contributions
Conceptualization, R.A.B, I.C.N and I.A.; methodology, R.A.B., I.C.N., J.L. and A.K.C.M.N.; software, J.L.; validation, J.L., N.S.B. and R.A.B.; formal analysis, J.L. and N.S.B.; investigation, J.L.; resources, R.A.B., I.C.N. and A.K.C.M.N.; data curation, J.L., N.S.B. and R.A.B.; writing—original draft preparation, J.L. and N.S.B., writing—review and editing, J.L., N.S.B. and R.A.B.; visualization, J.L.; supervision, N.S.B, A.K.C.M.N. and R.A.B.; project administration, R.A.B.; funding acquisition, R.A.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by USM Research University Grant, grant number (1001/PPSP/8012256).
Availability of data and materials
The data used and analyzed in this study are available from the corresponding author on request. This will require an ethical permit.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and approved by the ethical review and the Medical Research & Ethics Committee, Ministry of Health Malaysia (NMRR-19-399-45866) and USM Human Research Ethics Committee (USM/JEPeM/18010012).
Consent for publication
Informed consent was obtained from all subjects involved in the study.
Competing interests
The authors declare that they have no competing interests.
References
-
World Health Organisation. Hepatitis C [Internet]. 2022 [cited 2022 Sep 6]. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c.
.
-
Axley
P.,
Ahmed
Z.,
Ravi
S.,
Singal
A.K.,
Hepatitis C virus and hepatocellular carcinoma: a narrative review. Journal of clinical and translational hepatology.
2018;
6
(1)
:
79
.
View Article PubMed Google Scholar -
Chen
S.L.,
Morgan
T.R.,
The natural history of hepatitis C virus (HCV) infection. International Journal of Medical Sciences.
2006;
3
(2)
:
47-52
.
View Article PubMed Google Scholar -
European Assocication for the Study of Liver. EASL Clinical Practice Guidelines: Management of hepatitis C virus infection. Journal of Hepatology.
2011;
55
(2)
:
245-64
.
View Article PubMed Google Scholar -
Nouroz
F.,
Shaheen
S.,
Mujtaba
G.,
Noreen
S.,
An overview on hepatitis C virus genotypes and its control. The Egyptian Journal of Medical Human Genetics.
2015;
16
(4)
:
291-8
.
View Article Google Scholar -
P.V. Perumalswami,
R.S. Klein,
Hepatitis Viruses: Hepatitis C. In: Kaslow, R., Stanberry, L., Le Duc, J. (eds) Viral Infections of Humans. Springer, Boston, MA 2014.
View Article Google Scholar -
Wenger
P.J.,
Rottnek
F.,
Parker
T.,
Crippin
J.S.,
Assessment of Hepatitis C Risk Factors and Infection Prevalence in a Jail Population. Am J Public Health.
2014;
104
(9)
:
1722
.
-
Chan
D.P.,
Sun
H.Y.,
Wong
H.T.,
Lee
S.S.,
Hung
C.C.,
Sexually acquired hepatitis C virus infection: a review. International Journal of Infectious Diseases.
2016;
49
:
47-58
.
View Article PubMed Google Scholar -
Dwyre
D.M.,
Fernando
L.P.,
Holland
P.V.,
Hepatitis B, hepatitis C and HIV transfusion-transmitted infections in the 21st century. Vox Sanguinis.
2011;
100
(1)
:
92-8
.
View Article PubMed Google Scholar -
McCance-Katz
E.F.,
Valdiserri
R.O.,
Hepatitis C Virus Treatment and Injection Drug Users: It Is Time to Separate Fact From Fiction. Annals of internal medicine.
2015;
163
(3)
:
224-225
.
View Article Google Scholar -
Grebely
J.,
Prins
M.,
Hellard
M.,
Cox
A.L.,
Osburn
W.O.,
Lauer
G.,
Hepatitis C virus clearance, reinfection, and persistence, with insights from studies of injecting drug users: towards a vaccine. The Lancet infectious diseases.
2012;
12
(5)
:
408-414
.
View Article Google Scholar -
Degenhardt
L.,
Peacock
A.,
Colledge
S.,
Leung
J.,
Grebely
J.,
Vickerman
P.,
Global prevalence of injecting drug use and sociodemographic characteristics and prevalence of HIV, HBV, and HCV in people who inject drugs: a multistage systematic review. The Lancet. Global Health.
2017;
5
(12)
:
e1192-207
.
View Article PubMed Google Scholar -
Messina
J.P.,
Humphreys
I.,
Flaxman
A.,
Brown
A.,
Cooke
G.S.,
Pybus
O.G.,
Global distribution and prevalence of hepatitis C virus genotypes. Hepatology (Baltimore, Md.).
2015;
61
(1)
:
77-87
.
View Article PubMed Google Scholar -
Irekeola
A.A.,
Malek
N.A.,
Wada
Y.,
Mustaffa
N.,
Muhamad
N.I.,
Shueb
R.H.,
Prevalence of HCV genotypes and subtypes in Southeast Asia: A systematic review and meta-analysis. PLoS One.
2021;
16
(5)
:
e0251673
.
View Article PubMed Google Scholar -
Doan
T.Q.,
Hepatitis c in developing countries in Southeast Asia. InHepatitis C in developing countries 2018 Jan 1 (pp. 97-104). Academic Press..
.
-
Ho
S.H.,
Ng
K.P.,
Kaur
H.,
Goh
K.L.,
Genotype 3 is the predominant hepatitis C genotype in a multi-ethnic Asian population in Malaysia. Hepatobiliary & Pancreatic Diseases International.
2015;
14
(3)
:
281-6
.
View Article PubMed Google Scholar -
Mohamed
N.A.,
Rashid
Z.Z.,
Wong
K.K.,
Abdullah
S.A.,
Rahman
M.M.,
Hepatitis C genotype and associated risks factors of patients at University Kebangsaan Malaysia Medical Centre. Pakistan journal of medical sciences.
2013;
29
(5)
:
1142
.
View Article PubMed Google Scholar -
Elimination Of Mother-To-Child Transmission of HIV and Syphilis In Malaysia Towards Elimination Of Viral Hepatitis In Malaysia Through Multisectoral Collaboration, 2019 Case Study Ministry Of Health Malaysia.
.
-
Nishitsuji
H.,
Funami
K.,
Shimizu
Y.,
Ujino
S.,
Sugiyama
K.,
Seya
T.,
Hepatitis C virus infection induces inflammatory cytokines and chemokines mediated by the cross talk between hepatocytes and stellate cells. Journal of virology.
2013;
87
(14)
:
8169-8178
.
View Article PubMed Google Scholar -
Koziel
M.J.,
Cellular immune responses against hepatitis C virus. Clinical Infectious Diseases.
2005;
41
:
25-31
.
View Article PubMed Google Scholar -
Shin
E.C.,
Sung
P.S.,
Park
S.H.,
Immune responses and immunopathology in acute and chronic viral hepatitis. Nature Reviews Immunology.
2016;
16
(8)
:
509-523
.
View Article PubMed Google Scholar -
Chigbu
D.I.,
Loonawat
R.,
Sehgal
M.,
Patel
D.,
Jain
P.,
Hepatitis C Virus Infection: Host–Virus Interaction and Mechanisms of Viral Persistence. Cells.
2019;
8
(4)
:
376
.
View Article PubMed Google Scholar -
Heim
M.H.,
Innate immunity and HCV. Journal of Hepatology.
2013;
58
(3)
:
564-74
.
View Article PubMed Google Scholar -
Fallahi
P.,
Ferri
C.,
Ferrari
S.M.,
Corrado
A.,
Sansonno
D.,
Antonelli
A.,
Cytokines and HCV-related disorders. Clinical and Developmental Immunology.
2012;
2012
:
468107
.
View Article Google Scholar -
Nelson
J.E.,
Handa
P.,
Aouizerat
B.,
Wilson
L.,
Vemulakonda
L.A.,
Yeh
M.M.,
Network
NASH Clinical Research,
Increased parenchymal damage and steatohepatitis in Caucasian non-alcoholic fatty liver disease patients with common IL1B and IL6 polymorphisms. Alimentary Pharmacology & Therapeutics.
2016;
44
(11-12)
:
1253-64
.
View Article PubMed Google Scholar -
Murphy
C.,
de Chaumont
C.,
Shahnazaryan
D.,
Malone
C.,
Jefferies
C.,
Smith
S.,
Systemic IL-1β production as a consequence of corneal HSV-1 infection \textendash contribution to the development of Herpes Simplex Keratitis. Acta Ophthalmologica.
2015;
93
:
n-a
.
View Article Google Scholar -
Kasprzak
A.,
Zabel
M.,
Biczysko
W.,
Wysocki
J.,
Adamek
A.,
Spachacz
R.,
Expression of cytokines (TNF-α, IL-1α, and IL-2) in chronic hepatitis C: comparative hybridocytochemical and immunocytochemical study in children and adult patients. The Journal of Histochemistry and Cytochemistry.
2004;
52
(1)
:
29-38
.
View Article PubMed Google Scholar -
Behzadi
P.,
Sameer
A.S.,
Nissar
S.,
Banday
M.Z.,
Gajdács
M.,
García-Perdomo
H.A.,
The Interleukin-1 (IL-1) Superfamily Cytokines and Their Single Nucleotide Polymorphisms (SNPs). Journal of Immunology Research.
2022;
2022
:
2054431
.
View Article PubMed Google Scholar -
Shastry
B.S.,
SNPs: impact on gene function and phenotype. Methods in Molecular Biology (Clifton, N.J.).
2009;
578
:
3-22
.
View Article PubMed Google Scholar -
Ariffin
W.N.,
Sample size calculator [Internet]. 2022 [cited 2022 Sep 7]. Available from: https://wnarifin.github.io/ssc/ss2prop.html.
.
-
Vicknasingam
B.,
Narayanan
S.,
Navaratnam
V.,
Prevalence rates and risk factors for hepatitis C among drug users not in treatment in Malaysia. Drug and Alcohol Review.
2009;
28
(4)
:
447-54
.
View Article PubMed Google Scholar -
Pociot
F.,
M∅lvig
J.,
Wogensen
L.,
Worsaae
H.,
Nerup
J.,
A TaqI polymorphism in the human interleukin-1 β (IL-1 β) gene correlates with IL-1 β secretion in vitro. European Journal of Clinical Investigation.
1992;
22
(6)
:
396-402
.
View Article PubMed Google Scholar -
di Giovine
F.S.,
Takhsh
E.,
Blakemore
A.I.,
Duff
G.W.,
Single base polymorphism at -511 in the human interleukin-1 β gene (IL1 β). Human Molecular Genetics.
1992;
1
(6)
:
450-450
.
View Article PubMed Google Scholar -
Martinez
P.,
Lien
L.,
Zemore
S.,
Bramness
J.G.,
Neupane
S.P.,
Circulating cytokine levels are associated with symptoms of depression and anxiety among people with alcohol and drug use disorders. Journal of neuroimmunology.
2018;
318
:
80-86
.
View Article PubMed Google Scholar -
Lachance
J.,
. Encyclopedia of Evolutionary Biology.
2016;
2016
:
208-211
.
View Article Google Scholar -
Namipashaki
A.,
Razaghi-Moghadam
Z.,
Ansari-Pour
N.,
The essentiality of reporting Hardy-Weinberg equilibrium calculations in population-based genetic association studies. Cell Journal (Yakhteh).
2015;
17
(2)
:
187
.
View Article PubMed Google Scholar -
Wigginton
J.E.,
Cutler
D.J.,
Abecasis
G.R.,
A note on exact tests of Hardy-Weinberg equilibrium. The American Journal of Human Genetics.
2005;
76
(5)
:
887-893
.
View Article PubMed Google Scholar -
Omran
M.H.,
Ibrahim
N.E.,
Youssef
S.S.,
Fatouh
B.E.,
Nabil
W.,
El-Shami
M.M.,
Relation of interleukin-1β gene to treatment response in chronic patients infected with HCV genotype 4. Journal of Infection in Developing Countries.
2013;
7
(11)
:
851-8
.
View Article PubMed Google Scholar -
Ksiaa Cheikhrouhou
L.,
Sfar
I.,
Aounallah-Skhiri
H.,
Aouadi
H.,
Jendoubi-Ayed
S.,
Ben Abdallah
T.,
Cytokine and apoptosis gene polymorphisms influence the outcome of hepatitis C virus infection. Hepatobiliary {&}amp; Pancreatic Diseases International.
2011;
10
(3)
:
280-8
.
View Article PubMed Google Scholar -
Bahr
M.J.,
el Menuawy
M.,
Boeker
K.H.,
Musholt
P.B.,
Manns
M.P.,
Lichtinghagen
R.,
Cytokine gene polymorphisms and the susceptibility to liver cirrhosis in patients with chronic hepatitis C. Liver International.
2003;
23
(6)
:
420-5
.
View Article PubMed Google Scholar -
Minton
E.J.,
Smillie
D.,
Smith
P.,
Shipley
S.,
McKendrick
M.W.,
Gleeson
D.C.,
Trent Hepatitis C Study Group
Clearance of hepatitis C virus is not associated with single nucleotide polymorphisms in the IL-1, -6, or -10 genes. Human Immunology.
2005;
66
(2)
:
127-32
.
View Article PubMed Google Scholar -
Toro
D.M.,
Ramasawmy
R.,
Silva Neto
P.V.,
Pereira
G.L.,
Sarmento
P.S.,
Dray
H.L.,
Inflammasome genes polymorphisms may influence the development of hepatitis C in the Amazonas, Brazil. PLoS One.
2021;
16
(6)
:
e0253470
.
View Article PubMed Google Scholar -
Wang
Y.,
Kato
N.,
Hoshida
Y.,
Yoshida
H.,
Taniguchi
H.,
Goto
T.,
Interleukin-1β gene polymorphisms associated with hepatocellular carcinoma in hepatitis C virus infection. Hepatology (Baltimore, Md.).
2003;
37
(1)
:
65-71
.
View Article PubMed Google Scholar -
Tanaka
Y.,
Furuta
T.,
Suzuki
S.,
Orito
E.,
Yeo
A.E.,
Hirashima
N.,
Impact of interleukin-1β genetic polymorphisms on the development of hepatitis C virus-related hepatocellular carcinoma in Japan. The Journal of Infectious Diseases.
2003;
187
(11)
:
1822-5
.
View Article PubMed Google Scholar -
Shrivastava
S.,
Mukherjee
A.,
Ray
R.,
Ray
R.B.,
Hepatitis C virus induces interleukin-1β (IL-1β)/IL-18 in circulatory and resident liver macrophages. Journal of Virology.
2013;
87
(22)
:
12284-90
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 10 No 2 (2023)
Page No.: 5550-5557
Published on: 2023-02-28
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 4198 times
- PDF downloaded - 1263 times
- XML downloaded - 108 times
 Biomedpress
Biomedpress