Abstract
Early detection of relapse following successful curative-intent breast cancer surgery is an essential strategy for survival. Unfortunately, conventional screening via radiological imaging and tissue biopsies may be confounded by tumor size. Moreover, tissue biopsy is comparably invasive, clinically challenging, and predisposes medical and mental health complications. The use of liquid biopsies for postoperative screening in breast cancer survivors has been explored as a safer and easier alternative. The increasing popularity of the minimally invasive approach is due to its simplicity in accessing and obtaining the analyte and its lower associated costs. Using circulating tumor DNA (ctDNA) as a novel marker, the method enables real-time monitoring of disease-free survival and detection of potential cancer recurrence through cancer-specific alterations in ctDNA release from cancer cells and its short half-life. The promising clinical applications of ctDNA have fueled interest in developing new biomarkers with superior sensitivity for earlier detection of cancer recurrence, leading to the improved efficacy and efficiency of treatments and management. However, establishing this less invasive screening tool remains a challenge. In this review, we discuss the state-of-the-art in ctDNA-based liquid biopsies with a focus on the recent progress, challenges, and future directions of this technique for clinical applications in the follow-up care of breast cancer survivors.
INTRODUCTION
Breast cancer is one of the leading causes of death among women1, 2, 3. In 2020, the total number of new cases of breast cancer worldwide was estimated at 2.3 million1. For survivors who undergo primary curative-intent surgical resection, the goals of management are to achieve cancer-free survival and to prevent recurrence. Unfortunately, approximately 30% of these survivors develop recurrence after a primary surgical intervention, with about one-third of them subsequently succumbing to death4. Therefore, measures to confirm remission and to detect relapse at the earliest stage are the core strategies to improve prognosis.
Unfortunately, management goals are restricted by the limitations of conventional screening and diagnostic procedures. Tissue biopsy is the diagnostic gold standard and is used for all phases of diagnosis, staging, and prognosis. However, histological examination can be difficult to obtain due to challenges in retrieving the tumor tissue. Tissue biopsy is restricted by the invasive technique needed to access small tumor masses, which may lead to the acquisition of an inadequate sample from which to derive a conclusion. More importantly, when the tumor size is large enough to be detected to warrant a tissue biopsy, the recurrence has most likely metastasized to other parts of the body5, 6. Therefore, tissue biopsy is a comparably poor measure for the early detection of potential recurrence, impeding the timely evaluation of treatment options and the disease prognosis.
Unlike radiological screening, laboratory investigation remains crucial for diagnosis because breast cancer is highly heterogeneous, with diverse morphological features, molecular factors, and responses to therapy7, 8. At presentation, patients may be stratified to a specific treatment protocol based on empirical risk and the molecular subtype of cancer9, 10, 11. For example, anthracycline- (e.g., doxorubicin) and taxane-based agents (e.g., paclitaxel) are administered to patients with triple-negative breast cancer (TNBC) due to the lack of targetable receptors12, 13, 14, 15. Although these patients exhibit a positive response to therapy compared with non-TNBC patients, more than one-half of TNBC patients have cancer recurrence. These studies highlight that certain patients are not responsive to therapies, and many of them experience treatment-related adverse events. Furthermore, a growing body of evidence has demonstrated an association between therapy resistance and the development of recurrence16, 17, 18, 19. Therefore, parameters from laboratory investigations are fundamental for facilitating the precise planning, intervention, and subsequent follow-up regimens of postsurgical breast cancer patients.
Liquid biopsy has recently emerged as a promising new measure. Compared with conventional tissue biopsy, the practicality, cost efficiency, and the possibility to analyze a wide range of laboratory parameters have fueled interest in this new method. In this review, we will provide an overview of the clinical potential of liquid biopsy based on circulating tumor DNA (ctDNA) as a diagnostic and surveillance tool and highlight the challenges associated with ctDNA-based biomarkers, with an emphasis on perspectives from other solid tumors due to a lack of evidence in breast cancer.
THE CONCEPT OF LIQUID BIOPSY IN CANCER
Liquid biopsy offers a comprehensive view of the genomics of primary and metastatic tumors20, 21, 22, 23. This technique allows for the identification and screening of specific mutations in tumors using minimally invasive methods in real time. This is possible because circulating genetic materials derived from tumors can be found in biofluids, such as blood, urine, saliva, cerebrospinal fluid, pleural effusion, pericardial effusion, and ascites effusion24, 25, 26. Because it is possible for sampling to be performed without sedation in a routine clinic setting, liquid biopsy can be obtained during follow-up visits at any cancer stage or in the asymptomatic population5, 27. These properties allow for more regular, accessible, cost-efficient, and timely screening and surveillance that is comparable regarding quality of care for various members of the breast cancer survivor population.
Therefore, researchers may gain access to circulating genetic materials via liquid biopsy. The most common circulating genetic materials are exosomes, tumor-educated platelets, circulating tumor cells, microRNA, and cell-free DNA (cfDNA). In cancer, liquid biopsies are grouped based on the origin of tumor-derived materials, which include ctDNA, circulating tumor cells, tumor-derived exosomes, and other extracellular vehicles (EVs)6, 28. Early, practical, and regular access to these tumor-derived materials are the key advantages of liquid biopsy over other methods.
ctDNA
Biology of ctDNA
ctDNA may carry discriminatory information for screening and surveilling the presence of active cancer cells. ctDNA is a small fragment of cfDNA. It consists of fragments of double-stranded nuclear and mitochondrial DNA (mtDNA) approximately 40–200 base pairs (bp) in size24, 29. It has a peak at roughly 166 bp that corresponds to nucleosome-associated DNA fragments. A substantial number of studies have reported that ctDNA is shorter than cfDNA derived from noncancerous cells29, 30. Of interest, shorter fragments (<100 bp) might be enriched with ctDNA and mtDNA and may preferentially carry tumor-derived genomic alterations. The half-life of ctDNA is reported to be less than 2 hours; it is shorter than the half-life of any protein marker in the plasma, which can be several weeks31, 32. The short half-life of ctDNA makes it viable for the real-time monitoring of tumor burden in cancer patients. These distinguishing features of ctDNA enable researchers to infer the presence or absence of tumor residuals to deduce complete remission post-curative-intent multimodality treatments or to detect an early phase of recurrence.
Further pursuing this interest, numerous studies have investigated the primary mechanism of DNA release from a tumor; apoptosis, necrosis, pyroptosis, and senescence, among other mechanisms, have been suggested based on various research findings33, 34, 35, 36, 37. Short fragments of ctDNA are believed to be released into the blood or lymphatic circulation during apoptosis or phagocytosis by macrophages. This provides a clue as to why liquid biopsy can provide genetic information about the tumor. In addition, ctDNA can be released into the circulation by living tumor cells, either from primary or metastatic tumors, through EVs that play a role in the transportation of ctDNA between distant tissues. The size of EVs has a significant impact on the enrichment of ctDNA in tumor cells, as reported in prostate cancer patients24. More efforts are currently being invested to elucidate the components of ctDNA transported in EVs that are actively released by tumor cells and the effects of different treatments on this ctDNA. Therefore, research on the primary mechanism of DNA release from tumor cells may help to explain certain ctDNA parameters from liquid biopsies to facilitate evidence-informed clinical decision making.
Clinical applications of liquid biopsy-based ctDNA
Different ctDNA parameters from liquid biopsies may communicate the molecular events that underlie breast cancer pathogenesis. With optimal uses and interpretations of biomarkers, clinicians may formulate targeted interventions, monitor treatment response, and sensitively detect any residual active pathologies to address potential recurrence and metastasis. Unfortunately, in the case of tissue biopsy, the need to retrieve adequate tissue samples represents a major obstacle to achieving this goal. For example, it has been documented that epidermal growth factor receptor (EGFR) genetic testing is not conducted in approximately 19% of advanced non-small cell lung cancer (NSCLC) cases for the abovementioned reason28. In contrast, as ctDNA carries tumor-specific genetic and epigenetic alterations38, 39, 40; the abundance and easy access to samples in liquid biopsy make it a remarkable and practical alternative for the diagnosis and monitoring of cancer.
ctDNA as a prognostic marker
ctDNA that bears cancer-specific genetic and epigenetic aberrations has also been investigated as a feasible parameter for evaluating prognosis. Elevation of ctDNA in the circulation has been shown to positively correlate with a higher chance of survival. Patients who underwent resection for colorectal cancer shed high concentrations of preoperative ctDNA that correlated with cancer recurrence and poor survival outcomes29. In line with this finding, a prospective multicenter study involving 177 patients reported that elevated levels of ctDNA (defined as ≥5 ctDNA cells per 7.5 mL of blood) were associated with poor prognosis among patients with metastatic breast cancer41. These findings were further corroborated by a study by Parkinson and Gale42.
ctDNA as a biomarker for tumor burden
Tumor burden is another crucial aspect of breast cancer management. However, regular radiological assessment is limited by the risk of radiation exposure, and it is reliant on tumor size, which must be large enough to be detected by imaging modalities. In comparison, ctDNA can be used to evaluate tumor burden without these drawbacks. An advanced disease is postulated to shed a larger amount of ctDNA than a disease in its early stage or a premalignant lesion43, 44. In a retrospective study conducted by Parkinson and Gale42, ctDNA levels in serially collected liquid biopsy samples from 40 high-grade serous ovarian carcinomas were significantly correlated with lesion volume at the start of treatment. Likewise, Xu and Wei45 reported a significantly higher combined diagnostic score of ctDNA methylation markers in patients with residual tumors compared with patients with no detectable tumors. Of note, the diagnostic score varied between the early stages (I & II) and advanced stages (III & IV) of hepatocellular carcinoma. Therefore, the use of ctDNA to monitor tumor burden may be extended as a biomarker for cancer progression.
ctDNA as a heterogeneity marker
Cancer heterogeneity refers to variations in the characteristics and behavior of a tumor based on different types of cancer cells. Cancer heterogeneity presents a significant challenge to cancer diagnosis, survival prediction, treatment selection, and resistance46, 47, 48. Selective pressure toward a subpopulation within tumor cells may lead to acquired resistance mechanisms and subsequent recurrence47. The authors demonstrated that ctDNA may reveal mutations that were not seen in an archived biopsy as a result of either heterogeneity or de novo mutations47. Sequencing of ctDNA that bears specific genomic alterations can provide precise information about cancer heterogeneity to guide clinicians and oncologists in developing targeted interventions and optimal management plans.
ctDNA as a biomarker for therapeutic response
Response to treatment is the main focus of curative-intent breast cancer therapy. Easy access to liquid biopsy throughout the treatment period allows real-time monitoring of therapeutic response and disease progression. Chabon and Simmons49 demonstrated the utility of ctDNA in evaluating resistance mechanisms in 43 patients with NSCLC treated with the third-generation EGFR inhibitor rociletinib49. The findings suggest that the pattern of resistance following treatment with EGFR tyrosine kinase inhibitors is drug specific. Of note, type of therapy, radiation and chemotherapy dose, and exposure to the therapy can affect the ctDNA shedding mechanism and the level of ctDNA in the biofluids of cancer patients. Assessment of response to immunotherapy in NSCLC patients revealed high concordance between ctDNA and radiographic analyses. Interestingly, median time to initial therapy response measured using ctDNA was shorter than radiographic analysis (24.5 days vs 72.5 days, respectively), suggesting the highly sensitive clinical utility of ctDNA as a biomarker for therapeutic efficacy50. Furthermore, ctDNA levels in patients were used to assess the efficacy of combined therapies in the CheckMate 816 clinical trials. The study reported that ctDNA clearance was higher among patients who received neoadjuvant nivolumab plus chemotherapy than those who only received chemotherapy. Although more studies are required to establish the consistency of these findings, they indicate the promising utility of ctDNA as a surrogate marker of therapeutic efficacy.
ctDNA as a biomarker for monitoring minimal residual disease and recurrence 51, 52. For example, Kwok and Wu53 and Scherer and Kurtz54 reported that ctDNA was detectable in diffuse large B-cell lymphoma patients at the time of recurrence. Of note, ctDNA was detectable in minimal residual disease (MRD) before clinical recurrence in 73% of the patients. Another study by Reinert and Henriksen55 demonstrated early detection of MRD using longitudinal ctDNA data in patients with stage I–III colorectal cancer. Plasma analysis revealed that ctDNA-positive patients had remarkably shorter recurrence-free survival (RFS) compared with ctDNA-negative patients. A separate study reported shorter RFS in patients who received adjuvant chemotherapy with positive ctDNA status56. Increased ctDNA level in liquid biopsy has consistently been shown to be useful as an indicator of potential early recurrence.
CHALLENGES OF ctDNA-BASED BIOMARKERS
Despite the potential uses and applications of ctDNA, there are three key limitations to using ctDNA. The main challenge is that the method relies on a single type of analyte—ctDNA. Therefore, the amount of ctDNA, either in blood or urine, must be sufficient for analysis5, 6. In asymptomatic patients, a concentration of 1–10 ng/mL is required for the detection of ctDNA in a plasma sample34. A study by Haque and Elemento57 reported that a large volume of plasma is required to sufficiently detect the copy number of a ctDNA mutation in early-stage cancer. In breast cancer patients, a cutoff plasma DNA concentration was set at 120 ng/mL to achieve 100% sensitivity in early screening of the disease58. Achieving this sensitivity requires approximately 150–300 mL of blood sample per screening test. Furthermore, the percentage of ctDNA derived from cfDNA varies widely (0.01–90%), which complicates the mutational detection process. To circumvent the issue, technology platforms should adopt genome-wide technologies that can detect mutations at single loci with frequencies below 1% ctDNA25.
Second, for ctDNA to be utilized as a biomarker in clinical settings, the technology and technique for its detection and analysis must be accessible and practical to conduct in clinics or hospitals. Quantification or detection technologies, such as polymerase-chain reaction (PCR)-based methods (e.g. digital PCR, BEAMing) are highly sensitive and are able to detect ctDNA at very low allele frequencies (<1%); however, only specific and predetermined gene alterations can be detected26. In contrast, massively parallel sequencing, or next-generation sequencing, provides an extensive range of genetic alterations of the tumor. However, the technique requires dedicated bioinformaticians to analyze the results. Furthermore, the analysis must be carefully performed as low allele frequency variants are not distinguishable from background noise, which is often due to DNA polymerase errors.
Third, because the field is still growing, standard approaches to liquid biopsy have yet to be developed. These standards include methods for analyte extraction, confirmation of sample integrity, quantification, and analysis. For example, in sample processing, the presence of additional materials in serum derived from leukocyte lysis during clotting can dilute the ctDNA content25. The use of a stabilizer to increase the available blood processing window also requires attention, as certain components, such as heparin, can interfere with PCR processing activity. Moreover, techniques for processing plasma to extract plasma ctDNA, such as a magnetic bead, affinity column, and phenol-chloroform methods, vary in their ability to purify ctDNA. Consequently, the total quantity of isolated ctDNA can be skewed, leading to false negative or positive outcomes. Establishing standards for these aspects of liquid biopsy will ensure the quality of results to support valid interpretations and confidence in decision-making.
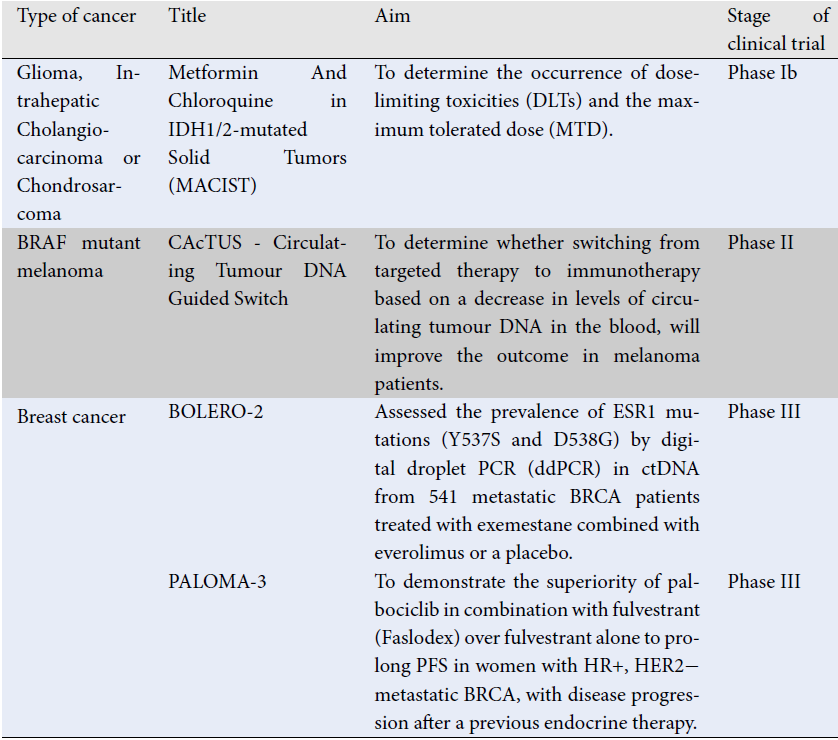
| Type of cancer | Title | Aim | Stage of clinical trial | Results | References |
|---|---|---|---|---|---|
| Glioma, Intrahepatic Cholangiocarcinoma or Chondrosarcoma | Metformin And Chloroquine in IDH1/2-mutated Solid Tumors (MACIST) | To determine the occurrence of dose-limiting toxicities (DLTs) and the maximum tolerated dose (MTD). | Phase Ib | Detection of tumor specific IDH1 hotspot mutations in ctDNA of patients with solid tumours treated with metformin and choloroquine. | NCT02496741 59 |
| BRAF mutant melanoma | CAcTUS - Circulating Tumour DNA Guided Switch | To determine whether switching from targeted therapy to immunotherapy based on a decrease in levels of circulating tumour DNA in the blood, will improve the outcome in melanoma patients. | Phase II | ctDNA decrease prompts switch to immunotherapy (nivolumab + ipilimumab) | NCT03808441 60 |
| Breast cancer | BOLERO-2 | Assessed the prevalence of ESR1 mutations (Y537S and D538G) by digital droplet PCR (ddPCR) in ctDNA from 541 metastatic BRCA patients treated with exemestane combined with everolimus or a placebo. | Phase III | Suggest that ESR1 mutations emerge in ctDNA from metastatic BRCA patients with prior aromatase inhibitors (AI) treatment, which can be detected by ddPCR, their presence is related with more aggressive tumours, and might be used as biomarkers for predicting outcome. | NCT00863655 61 |
| PALOMA-3 | To demonstrate the superiority of palbociclib in combination with fulvestrant (Faslodex) over fulvestrant alone to prolong PFS in women with HR+, HER2− metastatic BRCA, with disease progression after a previous endocrine therapy. | Phase III | Data have shown that patients with a high fraction of ctDNA have an overall worse PFS. | NCT01942135 62 | |
| Lung cancer | MILD | To measure the levels of cfDNA as a biomarker to assess whether this analysis can identify individuals at higher risk of cancer, improve the sensitivity and specificity of imaging techniques or both. | NA | The findings demonstrated that lung cancer, regardless of stage, is substantially related with high levels of cfDNA. The ctDNA level was unable to distinguish between healthy controls and patients with other tumours in small lung malignancies. ctDNA levels were found to be associated with disease aggression and a poor prognostic predictor for survival. For tumours in Stage II–IV, ctDNA levels tended to be significantly greater both at baseline and following surgery. | NCT02837809 63 |
| BENEFIT | To validate gefitinib response in lung adenocarcinomas bearing EGFR mutations | NA | imply that patients who would not benefit from EGFRTKI treatment can be identified through the analysis of EGFR mutation dynamics through ctDNA evaluation. | NCT02282267 64 | |
| Non-small cell lung cancer (NSCLC) | To assess dynamic changes in ctDNA levels in three treatment populations containing patients with NSCLC. | NA | ctDNA levels rise within hours to days of starting treatment. ctDNA testing during the acute post-treatment phase can reveal results that were not visible during the pre-treatment phase. ctDNA can be used as an alternative to tissue-based testing and can improve sensitivity for detecting treatment-resistant clones. | NCT03986463 65 | |
| Therapeutic Resistance and Clonal Evolution Assessed With Liquid Biopsy of NSCLC Patients in China | To evaluate the concordance of clonal mutations in ctDNA, using a 1021 gene targeted panel in plasma coupled with tumour tissue from patients with advanced-stage (IIB or IV) non-small-cell LC. | EGFR gene dominant clones were present in 72 patients, and multivariate analysis revealed that these clones constitute a distinct prognostic indication of the effectiveness of EGFR-TKI first-line therapy. Genomic data from tissue and ctDNA provide a more comprehensive picture of new and current actionable mutations that patients may benefit from additional therapeutic targets that may improve illness prognosis. | NCT03059641 66 | ||
| Colorectal cancer | AI-EMERGE | Focus on development of a non-invasive blood test for the early detection of cancer. Implemented automated machine learning (ML) to find and learn associations between cfDNA profiles and cancer ‘status’ to detect early-stage CRC. To evaluate the sensitivity and specificity of their test between CRC stages and compare them with current standard screening methods. | With 85% sensitivity and 85% specificity in CRC, it was shown that ML-based analysis is beneficial for determining the connection between a patient's cfDNA profile and cancer diagnosis. | NCT03688906 67 | |
| ECLIPSE (Evaluation of the ctDNA LUNAR test in an Average Patient Screening Episode) | To evaluate the performance characteristics of a blood-based ctDNA LUNAR-2 test to detect colorectal cancer in a screen-relevant, average-risk population. | Epigenomic technique dramatically improved ctDNA identification compared to somatic mutational analysis alone, with a 94% specificity for early-stage CRC patients (I-III). Future CRC incidence and mortality rates may be significantly affected by the usefulness of ctDNA testing as a non-invasive approach for early identification of CRC. | NCT04136002 68 |
FUTURE DIRECTIONS
Considerable progress has been made in the liquid biopsy field. There is increasing evidence to demonstrate the exemplary promise of ctDNA as a biomarker for the real-time monitoring of recurrence during the asymptomatic period. To provide a more precise estimation of recurrence risk, a better understanding of the origin and biology of ctDNA is needed. Further exploration of the association between ctDNA and cell apoptosis, necrosis, and shedding mechanisms may prove beneficial. To achieve this, more financial commitments, expertise sharing, and academic interest must be invested to advance our understanding of ctDNA release and clearance mechanisms for better interpretation of existing results, especially because ctDNA is already being used in clinical trials and clinical settings (Table 1). Studies in this direction may validate the consistency of findings. In addition, at the time of authoring this article, several clinical trials are being conducted to evaluate the clinical utility of ctDNA as a surrogate marker for treatment response, disease diagnosis, and risk of recurrence. Future randomized trials comparing ctDNA-guided decision-making to the standard of care may provide better insight into the utility of ctDNA in clinical oncology. Increasingly advanced genome sequencing technology for the analysis of ctDNA will highlight the applications of the liquid biopsy approach as an accurate and timely cancer biomarker.
Abbreviations
BC: Breast cancer, cfDNA: cell free DNA, ctDNA: Circulating tumor DNA, CTC: circulating tumor cells, EGFR: epidermal growth factor receptor, EVs: extracellular vehicles, TNBC: triple negative breast cancer, MRD: minimal residual disease, mtDNA: mitochondrial DNA, NSCLC: non-small cell lung cancer, RFS: recurrence-free survival
Acknowledgments
We thank Ministry of Higher Education (MOHE) Malaysia for financial support (FRGS/1/2020/SKK01/UNISZA/03/1).
Author’s contributions
Ramli RA contributed to conceptualization, data collection, writing-original draft preparation and review. Muhamad N and Ibrahim MS contributed to data collection, edited the first draft and review. All the authors potentially contributed, and approved the final version for publication.
Funding
Ministry of Higher Education (MOHE) Malaysia (FRGS/1/2020/SKK01/UNISZA/03/1).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Sung
H.,
Ferlay
J.,
Siegel
R.L.,
Laversanne
M.,
Soerjomataram
I.,
Jemal
A.,
Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians.
2021;
71
(3)
:
209-49
.
View Article PubMed Google Scholar -
Watkins
E.J.,
Overview of breast cancer. JAAPA: Official Journal of the American Academy of Physician Assistants.
2019;
32
(10)
:
13-7
.
View Article PubMed Google Scholar -
Ma
J.,
Jemal
A.,
Breast cancer statistics. Breast Cancer Metastasis and Drug Resistance: Progress and ProspectsSpringer New York, NY, USA 2013.
View Article Google Scholar -
Hiramanek
N.,
Breast cancer recurrence: follow up after treatment for primary breast cancer. Postgraduate Medical Journal.
2004;
80
(941)
:
172-6
.
View Article PubMed Google Scholar -
Chen
M.,
Zhao
H.,
Next-generation sequencing in liquid biopsy: cancer screening and early detection. Human Genomics.
2019;
13
(1)
:
34
.
View Article PubMed Google Scholar -
Yu
W.,
Hurley
J.,
Roberts
D.,
Chakrabortty
S.K.,
Enderle
D.,
Noerholm
M.,
Exosome-based liquid biopsies in cancer: opportunities and challenges. Annals of Oncology : Official Journal of the European Society for Medical Oncology.
2021;
32
(4)
:
466-77
.
View Article PubMed Google Scholar -
Rakha
E.A.,
Reis-Filho
J.S.,
Baehner
F.,
Dabbs
D.J.,
Decker
T.,
Eusebi
V.,
Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Research.
2010;
12
(4)
:
207
.
View Article PubMed Google Scholar -
Aleskandarany
M.A.,
Vandenberghe
M.E.,
Marchiò
C.,
Ellis
I.O.,
Sapino
A.,
Rakha
E.A.,
Tumour heterogeneity of breast cancer: from morphology to personalised medicine. Pathobiology.
2018;
85
(1-2)
:
23-34
.
View Article PubMed Google Scholar -
Polgár
C.,
Kahán
Z.,
Ivanov
O.,
Chorváth
M.,
Liga\vcová
A.,
Csejtei
A.,
Radiotherapy of Breast Cancer-Professional Guideline 1st Central-Eastern European Professional Consensus Statement on Breast Cancer. Pathology Oncology Research.
2022;
28
:
1610378
.
View Article PubMed Google Scholar -
Russnes
H.G.,
Lingj\aerde
O.C.,
B∅rresen-Dale
A.L.,
Caldas
C.,
Breast cancer molecular stratification: from intrinsic subtypes to integrative clusters. American Journal of Pathology.
2017;
187
(10)
:
2152-62
.
View Article PubMed Google Scholar -
Kaufmann
M.,
Pusztai
L.,
Members
B.E.,
Biedenkopf Expert Panel Members
Use of standard markers and incorporation of molecular markers into breast cancer therapy: Consensus recommendations from an International Expert Panel. Cancer.
2011;
117
(8)
:
1575-82
.
View Article PubMed Google Scholar -
Chen
S.Y.,
Tang
Y.,
Wang
S.L.,
Song
Y.W.,
Fang
H.,
Wang
J.Y.,
Timing of chemotherapy and radiotherapy following breast-conserving surgery for early-stage breast cancer: A retrospective analysis. Frontiers in Oncology.
2020;
10
:
571390
.
View Article PubMed Google Scholar -
Whelan
T.J.,
Olivotto
I.A.,
Parulekar
W.R.,
Ackerman
I.,
Chua
B.H.,
Nabid
A.,
Study Investigators
MA.20,
Regional nodal irradiation in early-stage breast cancer. The New England Journal of Medicine.
2015;
373
(4)
:
307-16
.
View Article PubMed Google Scholar -
Poortmans
P.M.,
Collette
S.,
Kirkove
C.,
Van Limbergen
E.,
Budach
V.,
Struikmans
H.,
Radiation Oncology
EORTC,
Breast Cancer Groups
Internal mammary and medial supraclavicular irradiation in breast cancer. The New England Journal of Medicine.
2015;
373
(4)
:
317-27
.
View Article PubMed Google Scholar -
Assidicky
R.,
Tokat
U.M.,
Tarman
I.O.,
Saatci
O.,
Ersan
P.G.,
Raza
U.,
Targeting HIF1-alpha/miR-326/ITGA5 axis potentiates chemotherapy response in triple-negative breast cancer. Breast Cancer Research and Treatment.
2022;
193
(2)
:
331-48
.
View Article PubMed Google Scholar -
Bai
X.,
Ni
J.,
Beretov
J.,
Graham
P.,
Li
Y.,
Cancer stem cell in breast cancer therapeutic resistance. Cancer Treatment Reviews.
2018;
69
:
152-63
.
View Article PubMed Google Scholar -
Saigusa
S.,
Tanaka
K.,
Toiyama
Y.,
Yokoe
T.,
Okugawa
Y.,
Ioue
Y.,
Correlation of CD133, OCT4, and SOX2 in rectal cancer and their association with distant recurrence after chemoradiotherapy. Annals of Surgical Oncology.
2009;
16
(12)
:
3488-98
.
View Article PubMed Google Scholar -
Kim
T.J.,
Lee
J.W.,
Song
S.Y.,
Choi
J.J.,
Choi
C.H.,
Kim
B.G.,
Increased expression of pAKT is associated with radiation resistance in cervical cancer. British Journal of Cancer.
2006;
94
(11)
:
1678-82
.
View Article PubMed Google Scholar -
Alharbi
M.,
Sharma
S.,
Guanzon
D.,
Lai
A.,
Zuñiga
F.,
Shiddiky
M.J.,
miRNa signature in small extracellular vesicles and their association with platinum resistance and cancer recurrence in ovarian cancer. Nanomedicine; Nanotechnology, Biology, and Medicine.
2020;
28
:
102207
.
View Article PubMed Google Scholar -
Lone
S.N.,
Nisar
S.,
Masoodi
T.,
Singh
M.,
Rizwan
A.,
Hashem
S.,
Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Molecular Cancer.
2022;
21
(1)
:
79
.
View Article PubMed Google Scholar -
Kruglyak
K.M.,
Chibuk
J.,
McLennan
L.,
Nakashe
P.,
Hernandez
G.E.,
Motalli-Pepio
R.,
Blood-based liquid biopsy for comprehensive cancer genomic profiling using next-generation sequencing: an emerging paradigm for non-invasive cancer detection and management in dogs. Frontiers in Veterinary Science.
2021;
8
:
704835
.
View Article PubMed Google Scholar -
Kavan
S.,
Kruse
T.A.,
Vogsen
M.,
Hildebrandt
M.G.,
Thomassen
M.,
Heterogeneity and tumor evolution reflected in liquid biopsy in metastatic breast cancer patients: a review. Cancer and Metastasis Reviews.
2022;
41
(2)
:
433-46
.
View Article PubMed Google Scholar -
Esposito
A.,
Criscitiello
C.,
Locatelli
M.,
Milano
M.,
Curigliano
G.,
Liquid biopsies for solid tumors: understanding tumor heterogeneity and real time monitoring of early resistance to targeted therapies. Pharmacology & Therapeutics.
2016;
157
:
120-4
.
View Article PubMed Google Scholar -
Sánchez-Herrero
E.,
Serna-Blasco
R.,
Robado de Lope
L.,
González-Rumayor
V.,
Romero
A.,
Provencio
M.,
Circulating Tumor DNA as a Cancer Biomarker: An Overview of Biological Features and Factors That may Impact on ctDNA Analysis. Frontiers in Oncology.
2022;
12
:
943253
.
View Article PubMed Google Scholar -
Donaldson
J.,
Park
B.H.,
Circulating tumor DNA: measurement and clinical utility. Annual Review of Medicine.
2018;
69
(1)
:
223-34
.
View Article PubMed Google Scholar -
Tay
T.K.,
Tan
P.H.,
Liquid biopsy in breast cancer: a focused review. Archives of Pathology & Laboratory Medicine.
2021;
145
(6)
:
678-86
.
View Article PubMed Google Scholar -
Bratulic
S.,
Gatto
F.,
Nielsen
J.,
The translational status of cancer liquid biopsies. Regenerative Engineering and Translational Medicine.
2021;
7
(3)
:
312-52
.
View Article Google Scholar -
Kwapisz
D.,
The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer?. Annals of Translational Medicine.
2017;
5
(3)
:
46
.
View Article PubMed Google Scholar -
Phallen
J.,
Sausen
M.,
Adleff
V.,
Leal
A.,
Hruban
C.,
White
J.,
Direct detection of early-stage cancers using circulating tumor DNA. Science Translational Medicine.
2017;
9
(403)
.
View Article PubMed Google Scholar -
Underhill
H.R.,
Kitzman
J.O.,
Hellwig
S.,
Welker
N.C.,
Daza
R.,
Baker
D.N.,
Fragment length of circulating tumor DNA. PLOS Genetics.
2016;
12
(7)
:
e1006162
.
View Article PubMed Google Scholar -
Duffy
M.J.,
Crown
J.,
Circulating Tumor DNA as a Biomarker for Monitoring Patients with Solid Cancers: Comparison with Standard Protein Biomarkers. Clinical Chemistry.
2022;
68
(11)
:
1381-90
.
View Article PubMed Google Scholar -
Chen
K.,
Zhao
H.,
Shi
Y.,
Yang
F.,
Wang
L.T.,
Kang
G.,
Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clinical Cancer Research.
2019;
25
(23)
:
7058-67
.
View Article PubMed Google Scholar -
Pessoa
L.S.,
Heringer
M.,
Ferrer
V.P.,
ctDNA as a cancer biomarker: A broad overview. Critical Reviews in Oncology/Hematology.
2020;
155
:
103109
.
View Article PubMed Google Scholar -
Wan
J.C.,
Massie
C.,
Garcia-Corbacho
J.,
Mouliere
F.,
Brenton
J.D.,
Caldas
C.,
Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nature Reviews. Cancer.
2017;
17
(4)
:
223-38
.
View Article PubMed Google Scholar -
Keller
L.,
Belloum
Y.,
Wikman
H.,
Pantel
K.,
Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. British Journal of Cancer.
2021;
124
(2)
:
345-58
.
View Article PubMed Google Scholar -
Schwarzenbach
H.,
Hoon
D.S.,
Pantel
K.,
Cell-free nucleic acids as biomarkers in cancer patients. Nature Reviews. Cancer.
2011;
11
(6)
:
426-37
.
View Article PubMed Google Scholar -
Kustanovich
A.,
Schwartz
R.,
Peretz
T.,
Grinshpun
A.,
Life and death of circulating cell-free DNA. Cancer Biology & Therapy.
2019;
20
(8)
:
1057-67
.
View Article PubMed Google Scholar -
Osumi
H.,
Shinozaki
E.,
Yamaguchi
K.,
Zembutsu
H.,
Clinical utility of circulating tumor DNA for colorectal cancer. Cancer Science.
2019;
110
(4)
:
1148-55
.
View Article PubMed Google Scholar -
Osumi
H.,
Shinozaki
E.,
Yamaguchi
K.,
Circulating tumor DNA as a novel biomarker optimizing chemotherapy for colorectal cancer. Cancers (Basel).
2020;
12
(6)
:
1566
.
View Article PubMed Google Scholar -
Zhang
L.,
Liang
Y.,
Li
S.,
Zeng
F.,
Meng
Y.,
Chen
Z.,
The interplay of circulating tumor DNA and chromatin modification, therapeutic resistance, and metastasis. Molecular Cancer.
2019;
18
(1)
:
36
.
View Article PubMed Google Scholar -
Cristofanilli
M.,
Budd
G.T.,
Ellis
M.J.,
Stopeck
A.,
Matera
J.,
Miller
M.C.,
Circulating tumor cells, disease progression, and survival in metastatic breast cancer. The New England Journal of Medicine.
2004;
351
(8)
:
781-91
.
View Article PubMed Google Scholar -
Parkinson
C.A.,
Gale
D.,
Piskorz
A.M.,
Biggs
H.,
Hodgkin
C.,
Addley
H.,
Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Medicine.
2016;
13
(12)
:
e1002198
.
View Article PubMed Google Scholar -
Allen
D.,
Butt
A.,
Cahill
D.,
Wheeler
M.,
Popert
R.,
Swaminathan
R.,
Role of cell-free plasma DNA as a diagnostic marker for prostate cancer. Annals of the New York Academy of Sciences.
2004;
1022
(1)
:
76-80
.
View Article PubMed Google Scholar -
Bettegowda
C.,
Sausen
M.,
Leary
R.J.,
Kinde
I.,
Wang
Y.,
Agrawal
N.,
Detection of circulating tumor DNA in early-and late-stage human malignancies. Science translational medicine.
2014;
6
(224)
:
224ra24-ra24
.
-
Xu
R.H.,
Wei
W.,
Krawczyk
M.,
Wang
W.,
Luo
H.,
Flagg
K.,
Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nature Materials.
2017;
16
(11)
:
1155-61
.
View Article PubMed Google Scholar -
Bedard
P.L.,
Hansen
A.R.,
Ratain
M.J.,
Siu
L.L.,
Tumour heterogeneity in the clinic. Nature.
2013;
501
(7467)
:
355-64
.
View Article PubMed Google Scholar -
El-Sayes
N.,
Vito
A.,
Mossman
K.,
Tumor heterogeneity: a great barrier in the age of cancer immunotherapy. Cancers (Basel).
2021;
13
(4)
:
806
.
View Article PubMed Google Scholar -
Crockford
A.,
Jamal-Hanjani
M.,
Hicks
J.,
Swanton
C.,
Implications of intratumour heterogeneity for treatment stratification. The Journal of Pathology.
2014;
232
(2)
:
264-73
.
View Article PubMed Google Scholar -
Chabon
J.J.,
Simmons
A.D.,
Lovejoy
A.F.,
Esfahani
M.S.,
Newman
A.M.,
Haringsma
H.J.,
Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nature Communications.
2016;
7
(1)
:
11815
.
View Article PubMed Google Scholar -
Goldberg
S.B.,
Narayan
A.,
Kole
A.J.,
Decker
R.H.,
Teysir
J.,
Carriero
N.J.,
Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clinical Cancer Research.
2018;
24
(8)
:
1872-80
.
View Article PubMed Google Scholar -
Tie
J.,
Wang
Y.,
Tomasetti
C.,
Li
L.,
Springer
S.,
Kinde
I.,
Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Science translational medicine.
2016;
8
(346)
:
346ra92-ra92
.
View Article Google Scholar -
Sch∅ler
L.V.,
Reinert
T.,
\Orntoft
M.W.,
Kassentoft
C.G.,
Árnadóttir
S.S.,
Vang
S.,
Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clinical Cancer Research.
2017;
23
(18)
:
5437-45
.
View Article PubMed Google Scholar -
Kwok
M.,
Wu
S.P.,
Mo
C.,
Summers
T.,
Roschewski
M.,
Circulating tumor DNA to monitor therapy for aggressive B-cell lymphomas. Current Treatment Options in Oncology.
2016;
17
(9)
:
47
.
View Article PubMed Google Scholar -
Scherer
F.,
Kurtz
D.M.,
Newman
A.M.,
Stehr
H.,
Craig
A.F.,
Esfahani
M.S.,
Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Science translational medicine.
2016;
8
(364)
:
364ra155-364ra155
.
View Article Google Scholar -
Reinert
T.,
Henriksen
T.V.,
Christensen
E.,
Sharma
S.,
Salari
R.,
Sethi
H.,
Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncology.
2019;
5
(8)
:
1124-31
.
View Article PubMed Google Scholar -
Tie
J.,
Cohen
J.D.,
Wang
Y.,
Li
L.,
Christie
M.,
Simons
K.,
Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut.
2019;
68
(4)
:
663-71
.
View Article PubMed Google Scholar -
Haque
I.S.,
Elemento
O.,
Challenges in using ctDNA to achieve early detection of cancer. BioRxiv.
2017;
:
237578
.
View Article Google Scholar -
Tangvarasittichai
O.,
Jaiwang
W.,
Tangvarasittichai
S.,
The plasma DNA concentration as a potential breast cancer screening marker. Indian Journal of Clinical Biochemistry.
2015;
30
(1)
:
55-8
.
View Article PubMed Google Scholar -
Raymond
V.M.,
Higashi
L.,
Marino
E.,
Lang
K.,
Evaluation of the ctDNA LUNAR-2 Test In an Average Patient Screening Episode (ECLIPSE). Journal of Clinical Oncology.
2021;
39
(no. 3_suppl)
:
1-1
.
View Article Google Scholar -
Khurshed
M.,
Molenaar
R.J.,
van Linde
M.E.,
Mathôt
R.A.,
Struys
E.A.,
van Wezel
T.,
A phase ib clinical trial of metformin and chloroquine in patients with idh1-mutated solid tumors. Cancers (Basel).
2021;
13
(10)
:
2474
.
View Article PubMed Google Scholar -
Lee
R.,
Rothwell
D.G.,
Chow
S.,
Shaw
H.M.,
Turajlic
S.,
Smith
N.,
Wolters Kluwer Health 2021.
Google Scholar -
Chandarlapaty
S.,
Chen
D.,
He
W.,
Sung
P.,
Samoila
A.,
You
D.,
Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncology.
2016;
2
(10)
:
1310-5
.
View Article PubMed Google Scholar -
O'Leary
B.,
Hrebien
S.,
Morden
J.P.,
Beaney
M.,
Fribbens
C.,
Huang
X.,
Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nature Communications.
2018;
9
(1)
:
896
.
View Article PubMed Google Scholar -
Pastorino
U.,
Silva
M.,
Sestini
S.,
Sabia
F.,
Boeri
M.,
Cantarutti
A.,
Prolonged lung cancer screening reduced 10-year mortality in the MILD trial: new confirmation of lung cancer screening efficacy. Annals of Oncology : Official Journal of the European Society for Medical Oncology.
2019;
30
(7)
:
1162-9
.
View Article PubMed Google Scholar -
Wang
Z.,
Cheng
Y.,
An
T.,
Gao
H.,
Wang
K.,
Zhou
Q.,
Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. The Lancet. Respiratory Medicine.
2018;
6
(9)
:
681-90
.
View Article PubMed Google Scholar -
Breadner
D.A.,
Vincent
M.D.,
Correa
R.,
Black
M.,
Warner
A.,
Sanatani
M.,
Exploitation of treatment induced tumor lysis to enhance the sensitivity of ctDNA analysis: A first-in-human pilot study. Lung Cancer (Amsterdam, Netherlands).
2022;
165
:
145-51
.
View Article PubMed Google Scholar -
Ai
X.,
Cui
J.,
Zhang
J.,
Chen
R.,
Lin
W.,
Xie
C.,
Clonal architecture of EGFR mutation predicts the efficacy of EGFR-tyrosine kinase inhibitors in advanced NSCLC: a prospective multicenter study (NCT03059641). Clinical Cancer Research.
2021;
27
(3)
:
704-12
.
View Article PubMed Google Scholar -
Putcha
Girish,
Liu
Tzu-Yu,
Ariazi
Eric,
Bertin
Marvin,
Drake
Adam,
Dzamba
Michael,
Hogan
Greg,
Kothen-Hill
Steven,
Liao
Jeffrey,
Li
Kang,
Blood-based detection of early-stage colorectal cancer using multiomics and machine learning. Abstract presented at: the American Society of Clinical Oncology (ASCO) GI Symposium.
2020;
:
23-25
.
Comments
Article Details
Volume & Issue : Vol 10 No 3 (2023)
Page No.: 5584-5595
Published on: 2023-03-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 9416 times
- PDF downloaded - 1429 times
- XML downloaded - 127 times
 Biomedpress
Biomedpress


