Abstract
Introduction: With the growing interest in exosomes for research and therapeutic applications, there's a critical need for effective enrichment strategies that can isolate exosomes in large quantities without contaminants. Despite various methods being developed, none have satisfactorily achieved this goal without contamination issues. This study introduces a novel approach for exosome separation - ion-exchange chromatography (IEC) - that leverages the negatively charged phospholipids on the exosomal surface for more effective isolation.
Methods: The study developed a novel exosome separation strategy using ion-exchange chromatography (IEC) and compared it with the traditional ultracentrifugation (UC) method. The comparison focused on yield, purity, and the biological effects of the exosomes enriched by both methods. The new IEC method was tested alongside classical separation techniques like ultrafiltration for its efficiency in large-scale exosome extraction.
Results: Both UC and IEC efficiently enriched exosomes with high abundance, but IEC was superior in terms of lower protein contamination and better particle dispersion. Furthermore, exosomes enriched with IEC exhibited a stronger clonogenic effect on murine lung epithelial cells compared to those enriched by UC. In wound healing assays, exosomes isolated by both IEC and UC significantly improved the healing ratio in lung epithelial cells, showcasing the potential therapeutic benefits of the exosomes isolated by these methods.
Conclusion: The novel ion-exchange chromatography (IEC) method developed for exosome separation demonstrated significant advantages in terms of yield, purity, and biological effects when compared to traditional ultracentrifugation. The lower protein contamination and better particle dispersion achieved with IEC, along with its superior clonogenic and healing effects on lung epithelial cells, suggest that IEC can play a crucial role in future exosome separation strategies, particularly for large-scale applications.
Introduction
Over the past few decades, exosomes have attracted considerable scientific interest1, 2, 3, 4. Most cells in all organisms secrete exosomes, which can load and transport specific proteins, RNA, and lipids to a target site for use, a key biological process5. Exosomes secreted by mesenchymal stem cells (MSCs) have been shown to exhibit broad therapeutic potential5, 6. Although therapeutic exosomes have application potential, challenges remain before they can be used in clinics2, 7, 8. One of the primary challenges lies in the necessity to prepare exosomes on a large scale, aiming to enhance production capacity and minimize batch variations when applied in commercial settings. Moreover, due to the impact of source cell condition and separation processes, there is a high susceptibility for biological activity deterioration during final utilization8. Therefore, it becomes crucial to maintain both morphology and activity of exosomes throughout the enrichment process. Additionally, controlling the overall extraction cost plays a vital role in expanding the range of commercial applications for exosomes. These prevailing circumstances pose novel tests for developing strategies that enable effective enrichment of exosomes.
Many techniques have been introduced for exosome isolation9. Ultracentrifugation, a technique commonly employed by researchers for exosome extraction without the need for special labeling, has been widely adopted in subsequent exosome studies10, 11. However, this method is time-consuming and expensive, and the resulting enriched exosomes often suffer from structural damage, clumping, and contamination with lipoproteins12, 13, 14. These drawbacks hinder the preservation of morphological integrity and biological activity of exosomes12, 15, 16. Microfluidic-based methods or immunoaffinity capture methods offer higher purity and lower contamination when isolating exosomes; thus, they are preferred for small sample purification in exosome diagnosis and purification. However, their high cost makes them unsuitable for large-scale sample purification. Size exclusion chromatography provides a feasible solution for large-scale separation of exosomes but faces challenges in distinguishing isolated exosomes from macromolecular proteins due to the principle of molecular sieve filtration. Therefore, it is crucial to develop an isolation technology that enables high-purity and large-scale preparation of exosomes.
Several anion-rich phospholipid molecules are present on the surface of exosomes. Therefore, it seems feasible to adsorb exosomes onto the fixed phase of liquid chromatography by negative and cation exchange and then elute and enrich the exosomes via ion re-exchange. The evident advantage offered by this approach is its ability to achieve large-scale exosome enrichment while preserving their morphology intact based on ion exchange principles, thereby facilitating optimal maintenance of their activity. In this study, we compared the newly developed ion exchange chromatography (IEC) with ultracentrifugation (UC) for isolating human MSC exosomes. Our goal was to develop and optimize separation methods that are more conducive to the promotion of exosome yield and purity. Furthermore, we aimed to prepare batches of homogeneous exosomes with stable effects at a lower cost, higher efficiency, and on a larger scale, to provide a solid foundation for exosome isolation, and to demonstrate their effects in clinical applications.
Methods
Cell Culture
Passage 1 human umbilical cord-derived mesenchymal stem cells (UC-MSCs) were obtained from Allcare Biomedical Development Co., LTD (Qingdao, China). The MSCs were cultured in Clin-SFM-Human MSC medium (1001A, Clin-Biotech, China) at 37 °C with 5% CO2. Upon reaching 60-80% confluency, MSCs were harvested using trypsin-EDTA (Gibco, Australia), counted, and then reseeded in T75 culture flasks at a density of 20,000 cells/cm2. In this experiment, cells from generations P6–P8 were used.
The mouse alveolar epithelial cell line (MLE-12) was purchased from ATCC and cultured in Dulbecco's Medium:Ham’s F12 (1:1) supplemented with 0.005 mg/mL insulin, 0.01 mg/mL transferrin, 30 nM sodium selenite, 10 nM hydrocortisone, 10 nM β-estradiol, 10 mM HEPES, 2 mM L-glutamine, 2% fetal bovine serum (Gibco), and 100 U/mL penicillin/streptomycin.
Separation of MSC Exosomes Using Ultracentrifugation and Ion-Exchange Chromatography
Ultracentrifugation
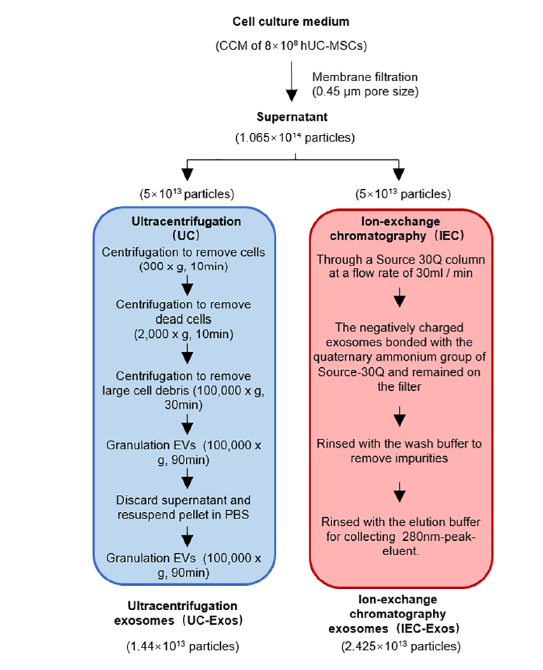
The 72-h MSC culture supernatant was collected as a raw material for exosome separation. The cell culture supernatant was centrifuged at 2000 ×g for 10 min to remove cells and large particle debris. The supernatant was then filtered through a 0.45 µm filter membrane and divided into two equal volumes for separation of exosomes using the Ion-Exchange Chromatography (IEC) method and the Ultracentrifugation (UC) method. The UC-Exos separation method, with slight modifications, followed Thery(9). Briefly, the treated cell supernatant was centrifuged at 100,000 ×g for 90 min. The exosome particles were resuspended in 10 mL phosphate-buffered saline (PBS) and centrifuged again at 100,000 ×g for 90 min. The resulting particles were resuspended in PBS, sub-packaged, and stored at −80 ℃ for subsequent steps.
Ion-Exchange Chromatography
The washing buffer consisted of 100 mM Tris-HCl, 10 mM EDTA, 0.4 M NaCl, pH 7.5. The elution buffer was composed of 100 mM Tris-HCl, 10 mM EDTA, 1 M NaCl, pH 7.5. The Source-30Q column was rinsed with wash buffer until the 280 nm detection line stabilized. A total of 750 mL of cell culture supernatant, filtered through a 0.45 µm filter, was loaded at a flow rate of 30 mL/min. During the loading process, the negatively charged exosomes bonded with the quaternary ammonium group of Source-30Q and remained on the filter. Impurities such as proteins not strongly electronegative were not retained and flowed out with the loading liquid. After sample loading, the column was rinsed with 1.5 Column Volumes (CV) of wash buffer to remove impurities, then switched to elution buffer for elution. In total, 200 mL of 280 nm-peak eluent was collected, sub-packaged, and stored at -80 °C for the next step. Both exosomes enriched by the UC protocol (UC_Exos) and those by the IEC protocol (IEC_Exos) were employed in this study. The workflow is shown in Figure 1.
| Volume (mL) | Particle population | Total particles | Yield (%) | |
|---|---|---|---|---|
| Original supernatant | 1500 | 7.1 × 10 10 | 1.065 × 10 14 | |
| UC | 24 | 6 × 10 11 | 1.44 × 10 13 | 28.8% |
| IEC | 250 | 9.7 × 10 10 | 2.425 × 10 13 | 48.5% |
| IEC + ultrafiltration | 120 | 2 × 10 11 | 2.4 × 10 13 | 48% |
Nanoparticle Tracking Analysis (NTA)
To determine particle size, potential, and concentration, NTA was performed using the ZetaView S/N 252 instrument (Particle Metrix, Inc., Mebane) equipped with ZetaView 8.04.02 SP2 software and a camera with an image scale of 0.703 µm/pixel. The exosomes were diluted in PBS at ratios ranging from 1:500 to 1:5000 to achieve a particle concentration of 107–108 particles/mL. Each sample was recorded in a 30-second video, three times.
Transmission Electron Microscopy (TEM)
TEM was utilized to observe the morphology of the exosomes and measure their size. Initially, the surface of the carbon film (Hatfield, PA) was made hydrophilic by glow discharge. After dilution with PBS, 5 µL of the exosome sample was placed on the carbon film and incubated for 2 min. Glutaraldehyde (2.5% w/v) was then added to 5 µL for 5 min. The carbon film was negatively stained with 5 µL 1% uranyl acetate for 1 min. Following these steps, the liquid was removed with a small piece of filter paper, and the exosomes on the carbon film were imaged using a Philips CM-100 TEM instrument.
Western Blotting
Several exosome markers were detected using western blotting. Exosomal proteins (5 µg) were electrophoresed and isolated on a 4%–20% SDS-PAGE gradient gel (Bio-Rad, Berkeley, CA, USA) and transferred to PVDF membranes. After blocking, the blot was incubated overnight with a 1:1000 dilution of each primary antibody at 4 °C. The membrane was then incubated with a horseradish peroxidase-conjugated secondary antibody for 1 h and detected using a chemiluminescence kit (Merck Millipore, Billerica, MA, USA).
CCK-8 Cell Viability Proliferation Test
The mouse alveolar epithelial cell line (MLE-12) was cultured in DMEM/F12 medium supplemented with 2% FBS and 0.01 mg/mL transferrin, and seeded in 96-well plates with 8000 cells/well and 100 µL culture medium. At a specified time point, 10 µL of CCK-8 solution (B34304, Bimake) was added to each well. The culture plate was returned to the incubator and incubated for 3 h, then the reaction signal at 450 nm was detected using an enzyme label reader.
Wound Healing Cell Scratch Test
MLE-12 cells were cultured in a six-well plate until confluency and then scratched with the tip of a sterile pipette to cause mechanical damage. The damaged monolayer of MLE-12 cells was washed three times with 1× PBS to remove cell debris and incubated at 37 ℃ and 5% CO2 for 24 h without exosome intervention. The scratched surface area was observed and photographed using an optical microscope (Olympus, Tokyo, Japan) at 0h, 6h, 12h, and 24h.
Clone Formation Assay
MLE-12 cells were seeded in a six-well plate at 700 cells/well. The culture was continued for 14 days or until the number of cells in most individual clones exceeded 50, during which the medium was changed every 3 days, and the cell status was observed. After cloning, cells were washed once with PBS, fixed with 1 mL of 4% paraformaldehyde per well for 30–60 min, washed again with PBS, and observed under an optical microscope.
Giemsa Staining
For adherent cultured cells that reached a specified time point, the medium was aspirated, and 4% paraformaldehyde (PFA) was added to fix the cell morphology. After 30 min, PFA was removed, and Giemsa staining solution (Solarbio, P1110) was added and incubated at room temperature for 30 min. Excess dye was washed off with PBS, and the sample was photographed under a light microscope.
Statistical Analysis
SPSS 20 was used for the statistical analysis of the data. All errors are presented as standard deviations. The two groups were compared using the Student’s t-test. Differences were considered statistically significant at P < 0.05.
Results
Isolation and Identification of Exosomes
NTA results showed that the average particle size of the exosomes isolated using UC and IEC was between 70 and 150 nm, respectively (Figure 2). The two exosomes with the same particle concentration were observed under TEM, showing that exosomes isolated using the two protocols exhibited a nearly circular vesicle structure (Figure 3A). Compared to exosomes separated via the UC protocol, those separated using IEC exhibited greater dispersion and a smaller average particle size, which may be related to the chromatographic separation process based on the charge adsorption principle. Long-term UC can easily aggregate exosome particles, making them difficult to disperse. However, exosome particles eluted from the packing adsorption column using the particle replacement principle were more dispersed, which may be more conducive to the diffusion of cell space and to the internalization and absorption by cells. Western blotting showed that the classical exosomal markers CD9, TSG101, and HSP70 were detected in both the source cells and the two types of exosomes (Figure 3B).
Comparison of the Extraction Efficiency of Exosomes Isolated by Two Methods
The extraction efficiency of exosomes using UC and IEC protocols was compared as shown in Table 1. The particle concentration of the sample was determined by NTA (Table 1). The starting material of each protocol was the MSC culture supernatant with a volume of 750 mL, and the measured particle concentration was 7.1 × 1010 particles/mL. Using the UC protocol, an exosome suspension was obtained with a volume of 24 mL and the concentration of particles measured was 6 × 1011 particles/mL. Using the IEC protocol, an exosome suspension with a volume of 250 mL was obtained and the concentration of particles measured was 9.7 × 1010 particles/mL.
The total number of particles is calculated according to the following formula:
The extraction efficiency of exosomes is calculated according to the following formula: Extraction efficiency = (total number of particles collected / total number of starting material particles) × 100%
Therefore, the extraction efficiency of the exosomes extracted using the UC protocol was 28.8%. The recovery rate of the exosomes extracted using the IEC protocol was 48.5%. Based on these results, the purification yield of the IEC protocol was significantly higher than that of the UC protocol. However, owing to the limitations of packing volume and eluent fixation, it is not possible to reach higher concentrations with less starting material per milliliter. Here, we attempted to combine the classical ultrafiltration protocol; that is, the exosomes collected using the IEC protocol were further concentrated through an ultrafiltration tube (Tiangen). This effectively compacted the exosome concentration to a particle count of 3.2 × 1013 in exosomes with a volume of 120 mL (recovery rate = 48%). Thus, a smaller amount of starting material can be used by IEC + ultrafiltration to obtain the same high recovery of exosome concentrates.
Comparison of Biological Effects of Exosomes Isolated by Two Methods
Exosomes promote the proliferation and migration of recipient cells through various mechanisms. To verify the difference between exosomes enriched by the UC protocol (UC_Exos) and exosomes enriched by the IEC protocol (IEC_Exos) in promoting the proliferation of receptor cells, the same amount of exosomes was co-cultured with mouse alveolar epithelial cells MLE-12 for 24, 48, 72, and 96 h. The final concentration of exosome protein in the medium was 50 µg/mL. At each time point, the absorbance at 450 nm was measured after incubation with the CCK-8 staining solution for 3 h. Figure 4A showed that the two types of exosomes exhibited significant proliferation-promoting effects in MLE-12 cells, although no statistically significant differences were detected between the two separation protocols. The clone formation assay (Figure 4B) showed that IEC_Exos promoted the formation of more clonal clumps in single cells than UC_Exos, suggesting that IEC_Exos may be more beneficial in promoting cell adhesion and proliferation. Furthermore, wound healing experiments were performed to compare the migration-promoting abilities of the two types of exosomes in recipient cells (Figure 5). At 0, 6, 12, and 24 h after scratching, the growth state of MLE-12 cells was photographed under an optical microscope. The results showed that the exosomes from both schemes had a better migration-promoting effect than the PBS group at each time point. This confirms that exosomes isolated using IEC exhibit the same biological activity as exosomes isolated by the classical UC protocol and even exhibit a stronger ability to promote cell adhesion in vitro.
Discussion
The potential application value of exosomes has increasingly garnered attention17, 18. Exosomes are heterogeneous in terms of their size, content, source, and function19. Thus, effective isolation and enrichment of exosomes will assist in evaluating their corresponding biological functions. The Ion Exchange Chromatography (IEC) method developed in this study employs liquid chromatography to separate exosomes. The anion groups on the surface of the stationary phase filler specifically bind to and replace the phospholipid molecules on the surface of the exosomes. As a result, the exosomes are retained in the stationary phase and enriched after a secondary replacement by a salt solution. The IEC method avoids the structural damage and clumping of exosomes caused by repeated centrifugation, unlike the Ultracentrifugation (UC) method. Notably, there is no clumping step in the entire separation process of IEC-exosomes, resulting in a homogeneous and well-separated exosome suspension after elution. Moreover, with the IEC method, the adsorption column retains only nucleic acids with an absorbance of about 280 nm and filters out proteins with an absorbance of about 320 nm. This significantly reduces the interfering protein content of exosomes in theory. More importantly, the IEC method is suitable for exosome separation in large samples.
A direct comparison between the newly developed IEC and the classical UC methods is crucial to establish an optimal method for isolating Mesenchymal Stem Cell (MSC) exosomes and to understand the properties of MSC exosomes. We compared the efficiency of the UC and IEC methods in isolating exosomes from the supernatant of mesenchymal stem cell cultures. Both UC and IEC protocols enriched exosomes abundantly, but the latter showed lower protein contamination and better particle dispersion. The biological effects of exosomes have been explored at the cellular level, including the well-documented effects of MSC-derived exosomes on lung epithelial proliferation and migration. Thus, we utilized the mouse alveolar epithelial cell line as a tool cell to verify the biological activities of MSC-derived exosomes. The findings demonstrated that exosomes enriched by UC and IEC significantly promoted proliferation in murine lung epithelial cells (MLE-12). Cloning experiments revealed that exosomes enriched by IEC exhibited a stronger clonogenic effect on lung epithelial cells. In wound healing experiments, exosomes enriched by both protocols significantly improved the lung epithelial healing ratio. These results confirm that exosomes isolated by UC and IEC exert positive biological effects in vitro.
We summarized the comparison between two enrichment strategies, UC and IEC. Firstly, compared with UC, IEC is not limited by equipment and is more suitable for separating exosomes from the supernatant of bulk cell cultures, offering advantages of ease and speed for commercial processes. Secondly, IEC does not affect exosome integrity like UC. Based on the principle of ion-exchange, vesicles with small charges are eluted along with RNA and proteins, leaving behind large-charged vesicles with good membrane integrity and uniform stability, which further improves the purity of exosomes. Thirdly, exosomes extracted by IEC are not precipitated or re-suspended as in UC, reducing the adhesion between exosomes and resulting in improved particle dispersion of the product. Fourthly, IEC can be easily combined with other separation methods to optimize exosomes, such as further concentration of enriched exosomes by combined ultrafiltration. Our findings show that the combined enrichment scheme of IEC and ultrafiltration is suitable for the isolation of exosomes from the supernatant of human umbilical cord mesenchymal stem cell cultures. Several protocols have been developed for using IEC for exosome separation and, combined with ultrafiltration, for replacing the solvent with a medium or PBS buffer suitable for in vivo experiments.
However, there are inevitable limitations to our strategy. Challenges in standardizing the procedure across different laboratories and ensuring reproducibility of results could pose limitations. Further harmonization of cell sources and standardized procedures should be undertaken to reduce these differences. In addition, more evaluation indicators of exosomes prepared by the IEC method need to be developed, such as the pro-biological effect of exosomes on various cells, the in vivo activity of exosomes, etc. The specificity of the technique for different exosome subpopulations might also be a limitation. The capacity of IEC to distinguish between exosomes with similar surface charges but different biological functions could be further explored. Evaluating the purity of isolated exosomes and the potential for non-exosomal RNA or protein contaminants remains a challenge.
Conclusions
This study developed a novel exosomal separation strategy, IEC, and combined it with classical exosome separation methods such as ultrafiltration, which showed significant advantages for large-scale extraction. Furthermore, through a comparison of in vitro cell experiments, the exosomes extracted by this protocol exhibited the same excellent bio-promoting activity as those extracted using the ultracentrifugation method. Therefore, IEC plays an important role in exosome separation strategies. When combined with several commonly used exosomal separation methods, it may be more conducive to promoting the yield and purity of exosomes. This approach aims to prepare batches of homogeneous exosomes with stable effects at lower cost, higher efficiency, and on a larger scale, demonstrating significant potential in clinical applications.
Abbreviations
CCK-8 - Cell Counting Kit-8, CV - Column Volumes, DMEM/F12 - Dulbecco's Modified Eagle Medium:Ham’s F12, FBS - Fetal Bovine Serum, IEC - Ion-Exchange Chromatography, MSC - Mesenchymal Stem Cells, NTA - Nanoparticle Tracking Analysis, PBS - Phosphate-Buffered Saline, PFA - Paraformaldehyde, PVDF - Polyvinylidene Difluoride, SDS-PAGE - Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis, SPSS - Statistical Package for the Social Sciences, TEM - Transmission Electron Microscopy, UC - Ultracentrifugation
Acknowledgments
None.
Author’s contributions
Conceptualization, Feng-Jun Xiao and Li-sheng Wang; Methodology and investigation, Xiaochen Cheng; Resources and data Curation, Li Du and Yuxin Lu; Original Draft Preparation, review and editing, Weiyuan Zhang; Funding Acquisition, Feng-Jun Xiao and Lisheng Wang. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Ha
D.H.,
Kim
H.K.,
Lee
J.,
Kwon
H.H.,
Park
G.H.,
Yang
S.H.,
Mesenchymal Stem/Stromal Cell-Derived Exosomes for Immunomodulatory Therapeutics and Skin Regeneration. Cells.
2020;
9
(5)
:
9
.
View Article PubMed Google Scholar -
Harrell
C.R.,
Jovicic
N.,
Djonov
V.,
Arsenijevic
N.,
Volarevic
V.,
Mesenchymal Stem Cell-Derived Exosomes and Other Extracellular Vesicles as New Remedies in the Therapy of Inflammatory Diseases. Cells.
2019;
8
(12)
:
8
.
View Article PubMed Google Scholar -
Zou
J.,
Yang
W.,
Cui
W.,
Li
C.,
Ma
C.,
Ji
X.,
Therapeutic potential and mechanisms of mesenchymal stem cell-derived exosomes as bioactive materials in tendon-bone healing. Journal of Nanobiotechnology.
2023;
21
(1)
:
14
.
View Article PubMed Google Scholar -
Lai
R.C.,
Tan
S.S.,
Yeo
R.W.,
Choo
A.B.,
Reiner
A.T.,
Su
Y.,
MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. Journal of Extracellular Vesicles.
2016;
5
(1)
:
29828
.
View Article PubMed Google Scholar -
Phinney
D.G.,
Pittenger
M.F.,
Concise Review: MSC-Derived Exosomes for Cell-Free Therapy. Stem Cells (Dayton, Ohio).
2017;
35
(4)
:
851-8
.
View Article PubMed Google Scholar -
Lotfy
A.,
AboQuella
N.M.,
Wang
H.,
Mesenchymal stromal/stem cell (MSC)-derived exosomes in clinical trials. Stem Cell Research & Therapy.
2023;
14
(1)
:
66
.
View Article PubMed Google Scholar -
Al-Khawaga
S.,
Abdelalim
E.M.,
Potential application of mesenchymal stem cells and their exosomes in lung injury: an emerging therapeutic option for COVID-19 patients. Stem Cell Research & Therapy.
2020;
11
(1)
:
437
.
View Article PubMed Google Scholar -
Doyle
L.M.,
Wang
M.Z.,
Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells.
2019;
8
(7)
:
8
.
View Article PubMed Google Scholar -
Zhang
Y.,
Bi
J.,
Huang
J.,
Tang
Y.,
Du
S.,
Li
P.,
Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. International Journal of Nanomedicine.
2020;
15
:
6917-34
.
View Article PubMed Google Scholar -
Zhu
L.,
Sun
H.T.,
Wang
S.,
Huang
S.L.,
Zheng
Y.,
Wang
C.Q.,
Isolation and characterization of exosomes for cancer research. Journal of Hematology & Oncology.
2020;
13
(1)
:
152
.
View Article PubMed Google Scholar -
Purushothaman
A.,
Exosomes from Cell Culture-Conditioned Medium: Isolation by Ultracentrifugation and Characterization. Methods in Molecular Biology (Clifton, N.J.).
2019;
1952
:
233-44
.
View Article PubMed Google Scholar -
Stam
J.,
Bartel
S.,
Bischoff
R.,
Wolters
J.C.,
Isolation of extracellular vesicles with combined enrichment methods. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences.
2021;
1169
.
View Article PubMed Google Scholar -
Baranyai
T.,
Herczeg
K.,
Onódi
Z.,
Voszka
I.,
Módos
K.,
Marton
N.,
Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS One.
2015;
10
(12)
.
View Article PubMed Google Scholar -
Coughlan
C.,
Bruce
K.D.,
Burgy
O.,
Boyd
T.D.,
Michel
C.R.,
Garcia-Perez
J.E.,
Exosome Isolation by Ultracentrifugation and Precipitation and Techniques for Downstream Analyses. Current Protocols in Cell Biology.
2020;
88
(1)
.
View Article PubMed Google Scholar -
Johnstone
R.M.,
Adam
M.,
Hammond
J.R.,
Orr
L.,
Turbide
C.,
Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). The Journal of Biological Chemistry.
1987;
262
(19)
:
9412-20
.
View Article PubMed Google Scholar -
An
M.,
Wu
J.,
Zhu
J.,
Lubman
D.M.,
Comparison of an Optimized Ultracentrifugation Method versus Size-Exclusion Chromatography for Isolation of Exosomes from Human Serum. Journal of Proteome Research.
2018;
17
(10)
:
3599-605
.
View Article PubMed Google Scholar -
Zhang
H.,
Zang
C.,
Zhao
W.,
Zhang
L.,
Liu
R.,
Feng
Z.,
Exosome Derived from Mesenchymal Stem Cells Alleviates Hypertrophic Scar by Inhibiting the Fibroblasts via TNFSF-13/HSPG2 Signaling Pathway. International Journal of Nanomedicine.
2023;
18
:
7047-63
.
View Article PubMed Google Scholar -
Wang
S.,
Lei
B.,
Zhang
E.,
Gong
P.,
Gu
J.,
He
L.,
Targeted Therapy for Inflammatory Diseases with Mesenchymal Stem Cells and Their Derived Exosomes: From Basic to Clinics. International Journal of Nanomedicine.
2022;
17
:
1757-81
.
View Article PubMed Google Scholar -
Tertel
T.,
Dittrich
R.,
Arsène
P.,
Jensen
A.,
Giebel
B.,
EV products obtained from iPSC-derived MSCs show batch-to-batch variations in their ability to modulate allogeneic immune responses in vitro. Frontiers in Cell and Developmental Biology.
2023;
11
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 11 No 4 (2024)
Page No.: 6348-6356
Published on: 2024-04-30
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 3738 times
- PDF downloaded - 1185 times
- XML downloaded - 74 times
 Biomedpress
Biomedpress






