Abstract
There is a broad spectrum of hematological diseases, and their origins can be attributed to a variety of factors, including genetic abnormalities such as leukemia, sickle cell anemia (SCA), and thalassemia, as well as conditions associated with the lack of certain blood components, such as iron deficiency anemia (IDA). Testing and analyzing for hematological disorders is intensive in terms of time, effort, and labor. Additionally, there is a higher chance of human error and variance during the manual examination and analysis of the test samples, depending on the expertise, skills, and experience of the examiner. Recent developments in artificial intelligence (AI), such as Machine Learning (ML) and Deep Learning (DL) algorithms—such as Random Forest (RF), Decision Tree (DT), and Support Vector Machine (SVM)—have demonstrated the considerable contribution they could make to more rapid and accurate disease diagnosis, detection, and classification. An increasing number of hematological diseases are being diagnosed using AI techniques, which combine tabular and image data to eliminate human error, generate more precise results, and decrease the time required for diagnosis. This review discusses several widely utilized AI disease evaluation algorithms and their applicability to hematological disorders. Additionally, we highlight key challenges such as the lack of accessible clinical data, which inhibits the implementation of AI in the field of medicine.
Introduction
An accurate early diagnosis is a crucial factor that plays a vital role in determining the treatment and prognosis for any disease, particularly in hematological disorders. In addition to the complete blood count (CBC), bone marrow (BM) and peripheral blood smears (PBS) examined under a microscope are two of the main tests used to diagnose a hematological disorder. However, the results and analysis of these diagnostic tests can vary based on the expertise and interpretation of the examiner. Hence, there is a high probability of significant human error, which could prove detrimental to a patient’s treatment plan and prognosis1.
Moreover, the time-intensive and labor-intensive nature of these tests adds to the testing costs, making them less accessible to a large portion of the population. For example, iron deficiency anemia (IDA) and thalassemia exhibit similar symptoms; therefore, when a misdiagnosis occurs, a thalassemic patient could be prescribed unnecessary iron supplements while the thalassemia could go untreated2. In the case of leukemia, a delayed diagnosis may result in rapid disease progression and possible death of the patient. Therefore, a correct diagnosis made at an early stage of the development of a hematological disorder is crucial as it enables medical professionals to develop suitable treatment plans. Automated approaches, such as artificial intelligence (AI), have been proposed as more accurate, time-efficient, and less labor-intensive means for disease detection1, 2, 3.
The early foundations of AI were laid in the 1970s with the introduction of rule-based systems like MYCIN, which were utilized for diagnosing bacterial infections and prescribing appropriate antibiotics4. Since the inception of AI in healthcare, it has advanced significantly and shown high success rates when utilized in numerous intelligent applications to solve several data-related issues5. A growing number of AI applications, and its subcategory Machine Learning (ML), are being developed in the field of medicine to assist clinicians in analyzing test results for rapid disease diagnosis and to devise better therapeutic interventions6, 7. This development was catalyzed by the abundance of clinical data combined with powerful AI tools, which have allowed for a wider range of applications in this area8.
The purpose of this review is to highlight the development of AI technology and its application in hematological diseases, as compared to other diseases like brain tumors where the field has been explored in detail. Additionally, with the recent exponential advancements in this technology, there are opportunities to implement a myriad of AI technologies. For example, analyzing blood cell morphology can be quite complex and variable, but it is becoming consistently more accurate and faster thanks to numerous AI algorithms and the availability of big data sets. Therefore, in this review, we discuss various models and examples of AI that are being applied in the field of medicine, particularly in hematological disorders. Additionally, the challenges faced in implementing AI in disease detection and classification are also discussed in this study.
Adjustments include spelling changes (e.g., "hematological" instead of "haematological") for consistency and readability, the use of semicolons to connect related clauses, and slight wording changes for clarity and flow.
Methods
This review screened three databases (PubMed, Google Scholar, and ScienceDirect) for relevant literature related to the term(s). Studies that utilized AI in the field of hematology and hematological cancers were searched using keywords like artificial intelligence, clinical translation, deep learning, hematological disorders, machine learning, diagnostics, healthcare technology, leukemia, anemia, and other terms related to artificial intelligence and its applications in the medical domain.
Machine Learning (ML)
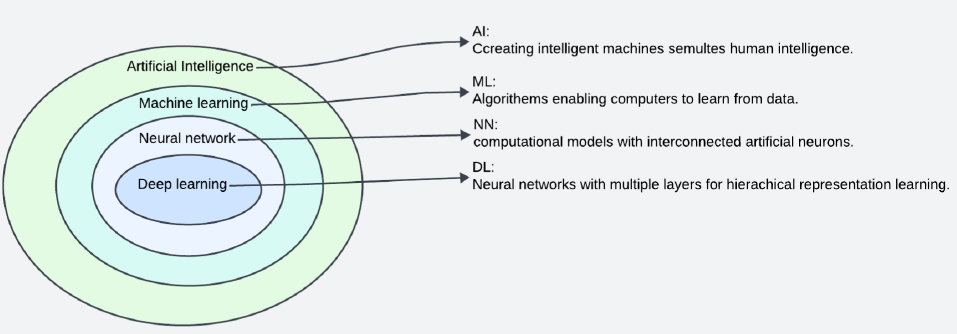
ML is a part of AI that analyzes data samples to create models using mathematical and statistical approaches, allowing machines to learn without programming (Figure 1 ). When a computer program is given a set of tasks, it is said to have learned from its experience if its quantifiable performance on those tasks improves over time as it completes them9. ML was first used in a checkers game by Arthur Samuel in 1959, who utilized annotated moves from experienced players10. Since then, the algorithm has been validated and applied to a wide range of different applications1, 10, 11, 12. Currently, with the rapid increase in computational capabilities and data availability, training data-driven ML models has become more feasible, resulting in more time and cost-effective ML applications13.
ML is classified mainly into two types based on the type of input: supervised and unsupervised. The supervised learning ML algorithm makes use of “labeled” input and output training datasets to learn from in order to classify unlabeled datasets and predict the output accurately once the supervised ML algorithm is refined14. On the other hand, unsupervised ML examines unlabeled datasets and reveals unknown patterns using various clustering and/or association algorithms14. Supervised ML is grouped into two subtypes based on the type of output: classification and regression15. Classification is used to classify datasets into specific segments based on chosen parameters, while regression uses statistical methods to find a correlation between dependent and independent variables, which helps to make the cause-and-effect prediction. The following sections provide a bird’s eye view of the frequently used supervised ML algorithms utilized in the medical field due to the homogeneous and consistent nature of clinical tests along with the availability of sufficient data to form training datasets.
Decision Tree (DT)
The modern DT algorithm was developed in 1986 by John Quinlan, who developed features from a family and its members addressing the same task16. According to Suthaharan (2016), this supervised learning method could be interpreted as a “hierarchical domain division technique” due to its role in splitting the data domain, also known as a “node.” Ideally, an optimized DT model would split the dataset into subsets leading to maximum information gain, hence leading to better classification17. To support this hierarchical structure, DT comprises leaf nodes, branches, and a root node. Every internal node contains an attribute test, followed by branching of the test result, and finally, leaf nodes that indicate class labels18. In a small dataset, DT is simple to interpret and the accuracy of DT is comparable to other classification algorithms; however, this could lead to overfit classification (Table 1).
| Algorithm | Advantages | Disadvantages |
| Decision tree (DT) 19 | •Small-sized trees are simple to interpret. | Over fit with classification in some dataset |
| Random forest (RF) 19 | •Managed unequal data sets with missing variables. | Over fit for some datasets with noisy classification task. |
| Support vector machine (SVM) 19 | •Works well in high dimensional space. | •Requires a long time for training with a huge data set. |
| K-nearest neighbor (KNN) 20 | •Implementation is simple and clear. | •Sensitivity to irrelevant or noisy data. |
| Naïve Bayes (NB) 21 | For classification, only a little quantity of training data is required. | Interaction between features cannot be learned because of feature independence. |
| Convolutional Neural Network (CNN) 22 | •It is used in image processing without the need for feature engineering. | •Longer time for training set |
Random Forest (RF)
The RF algorithm, initially introduced by Breiman in 2001, comprises a collection of predictive trees that are aggregated such that each tree is constructed using randomly selected vector values and sampled simultaneously while adhering to the same distribution23, 24. During the process, the prediction that acquires most of the tree votes will be predicted as the main result. RF possesses the capability to handle datasets containing missing variables and uneven data. Additionally, it is capable of computing crucial features for classification, which establishes it as one of the most efficient classifier algorithms in the domain (Table 1). It is widely used in detecting and classifying cancer cells, for example, in the detection of acute lymphoblastic leukemia (ALL)25, 26, 27, 28.
Support Vector Machine (SVM)
The SVM algorithm, first introduced in 1992 by Vladimir Vapnik, is utilized to determine a suitable class label to segment data samples29. SVM, in brief, is a learning algorithm that analyzes data used for categorization; when data patterns are highly dimensional and well-spaced, it is an excellent choice for classification30. With larger datasets, however, this approach requires an extended training period (Table 1). In addition to disease diagnosis, SVM is applied in the classification of hematological malignancies and their subtypes utilizing its categorization capabilities1, 31, 32.
K-Nearest Neighbor (KNN)
KNN is a non-parametric classification algorithm developed by Joseph Hodges and Evelyn Fix in 195133. The model attempts to classify non-linear sample data points in a database into several classes34. As a result, it tends to identify irrelevant data that are distant, so it takes a significantly longer time in the testing procedure (Table 1). In the medical and health industry, this method is used on electrocardiogram (ECG) data to detect abnormalities in the heart, such as tachycardia35.
Naïve Bayes (NB)
NB is a classification method based on Bayes' Theorem, developed by Thomas Bayes in 176036. It is an independent predictor hypothesis that evaluates the degree of relationship between the classification variables. Essentially, the class with the greatest likelihood is the class with the highest probability. It is effective with small datasets and has been implemented in the classification of hematological disorders and heart disease, among other disease diagnoses37, 38, 39. For instance, in the study of cardiovascular disease, scientists utilized NB as a data mining technique to discover correlations in a database between variables such as age, sex, and fasting blood sugar37, 40. NB is also applied in the classification of red blood cells (RBCs) and sickle cells39.
Deep Learning (DL)
DL is a sub-domain of ML comprised of numerous layers that extract information from various input data formats, including images, numbers, etc. The DL model derives its structure and operations from the neural network found in the human brain and thus closely resembles them41. The structure of its processing units consists of output, hidden, and input layers. Employing units or nodes, each layer's nodes are connected to those in the layer beneath it; each link is assigned a weight42. DL is divided into three subtypes: supervised, semi-supervised, and unsupervised. The main DL models that are currently being practiced are artificial neural networks (ANN), deep neural networks (DNN), recurrent neural networks (RNN), and convolutional neural networks (CNN). Leukemia31, malaria43, and thalassemia19 are among the many diseases that have been diagnosed using CNN in medical image processing. A key advantage of DL compared to conventional ML is that it eliminates the necessity for segmentation and feature extraction operations that are intrinsic to ML’s functionality.
Convolutional Neural Network (CNN)
CNN, which Yann LeCun introduced in 1980 as a subfield of artificial neural networks (ANN), is widely employed in the domain of image processing3, 20. CNN operates by assigning weights to the components within an image and then differentiating them from the remainder of the image; this makes it faster in the testing set than any other method (Table 1 ). Nonetheless, in applications where data is scarce, the algorithm's utility is restricted due to the necessity for sizable datasets to ensure greater accuracy and efficiency (Table 1). Over the past three decades, a specific type of CNN has proven effective in diagnosing cancer, particularly leukemia12.
Performance Metrics in AI Application
Performance metrics are crucial for evaluating the effectiveness of AI models, especially in the medical field, where accuracy is paramount. In this context, these metrics play a vital role in the AI development pipeline. AI models that achieve over 90% in these evaluations are typically regarded as highly effective or reliable, given the high stakes involved in patient care and diagnosis. The metrics that are commonly used are accuracy, sensitivity/recall, specificity, F1-Score, and precision21.
Accuracy: Accuracy is described as the proportion of true predictions made by a classifier to the total of all predictions as Equation 1 :
Sensitivity/Recall: Sensitivity gives only true positive measure considering total prediction and can be measured as Equation 2 :
Specificity: Specificity calculates how many true negatives are correctly detected and identified as Equation 3 :
F1 Score: The F1 score is the harmonic mean of accuracy and recall. Its calculation is as Equation 4 :
Precision: Precision is defined as the metric that measures the accuracy of predicted positive observations to all expected positive observations. It is determined as Equation 5 :
Application of AI in the Diagnosis of Hematological Disorders
In the area of hematology, a precise and rapid diagnosis of disease is necessary, as some blood disorders, such as acute myeloid leukemia (AML), are highly heterogeneous and exhibit transcriptomic, proteomic, and metabolomic variations22, 44, 45. Furthermore, depending on the knowledge and expertise of the hematologist, there is a significant chance of human error, which may delay or result in an incorrect diagnosis of the disease, causing severe disease progression or death. Thus, there is a growing need for a medical system that can reduce diagnosis turnaround time while preventing human errors.
In 1995, AI was first applied in a hematology laboratory for peripheral blood interpretation, flow cytometry immunophenotyping, and bone marrow reporting, and it became a strong point due to its high accuracy and specificity46. The implementation of AI in hematology includes, but is not limited to, diagnosis, detection, and classification of diseases. Among the most common uses for machine learning (ML) in hematology are image processing, recognition, and classification8. Moreover, ML and deep learning (DL) have been applied to the detection of diseases utilizing visual and tabular data, among others47. Visual data contains images of blood and bone marrow smears, while tabular data provides information such as age, gender, and test results.
| Author | Year | Disease | Classifier | Dataset | Performance metrics |
| Dasariragu et al . 27 | 2020 | AML | RF | Images of PBS | 92.99% accuracy for detection 93.45 accuracy for immature classification |
| Dese et al . 32 | 2021 | Leukaemia | SVM | Images of PBS | Accuracy ALL-100% AML-93.33% CLL-100% CML-93.33% |
| Kimura et al . 48 | 2019 | MDS, AA | CNN | Images of PBS | Sensitivity-96.2% Specificity-100% |
| Wang et al . 49 | 2022 | MDS, AA, AML | CNN | Images of BM | Accuracy-91.4% for MDS 92.9% accuracy for MDS, AA, AML |
Haematological Malignancies
Leukaemia
Leukaemia is a type of blood cancer characterized by the uncontrolled proliferation of immature white blood cells (WBCs), called blasts. It is divided into acute or chronic leukaemia based on the progression of the disease. Acute leukaemia is an aggressive disease with rapid development due to a sudden increase in blast cells, which may result in the death of patients if it is not treated at an early stage. In contrast, chronic leukaemia usually manifests at a slower rate32. Furthermore, leukaemia can be myeloid or lymphoid in origin, depending on the kind of cell afflicted by the disease. Therefore, leukaemia is separated into four types: Acute Lymphoid Leukaemia (ALL), Chronic Lymphoid Leukaemia (CLL), Acute Myeloid Leukaemia (AML), and Chronic Myeloid Leukaemia (CML)50.
Leukaemia is clinically diagnosed by examining cells from blood smears under a microscope. This procedure is time-consuming and may be subject to human error. AI solutions have addressed these issues by recognizing blast cells and helping pathologists and haematologists make decisions (Table 2)50. For instance, Dasariragu et al. (2020) proposed ML for the detection and classification of AML cells by using RF to segment the nucleus and cytoplasm based on morphological features. They extracted sixteen features for each image of WBCs from the blood smear sample. The RF algorithm was applied for the detection of immature cells and classification into four types of cells: erythroblasts, monoblasts, promyelocytes, and myeloblasts. The proposed method achieved 92.99% accuracy for detection and 93.45% for the classification of immature WBCs27.
Another study classified AML subtypes, M1 and M2, in line with the French-American-British (FAB) classification system by constructing an ML model51. Thirty-one AML-M1 and nineteen AML-M2 bone marrow smear images were analyzed for variables, of which two morphological, six radiomics, and one clinical feature became part of the four classification models. The random forest model and broad learning system (BLS) were among the four other classifiers (Naive Bayes, KNN, SVM, ANN) used in this study, which found the random forest model with all nine variables performed the best, as shown by the average accuracy (0.998 ± 0.003), AUC (0.998 ± 0.004), F1-score (0.998 ± 0.004), recall (0.996 ± 0.009), and precision (1 ± 0) of the model12.
Furthermore, Dese et al. (2021) suggested ML for all leukaemia types (ALL, CLL, AML, CML) by examining 250 images of blood smear slides that were taken from clinical samples. First, the KNN algorithm was used to segment WBCs by distinguishing them from RBCs and platelets, followed by the watershed method to separate overlapped cells. Next, the researchers used SVM for classification, whereby the combination of these two algorithms achieved the accuracy of 100%, 93.33%, 100%, 93.33%, and 100% in classifying ALL, AML, CLL, CML, and normal cell types, respectively32.
Multiple other investigations utilizing AI have also been able to differentiate leukaemia from other diseases. For example, Kimura et al. (2019) investigated the CNN method by employing extreme gradient boosting (XGBoost) to differentiate myelodysplastic syndrome (MDS) from aplastic anaemia (AA) with a sensitivity of 96.2% and a specificity of 100%48. Another study employed bone marrow smear pictures analyzed using the DL technique to differentiate between AML, MDS, and AA49. In the same investigation, the group classified MDS from non-MDS with 91.4% accuracy using the CNN recognition model; they distinguished AA, MDS, and AML with 92.9% accuracy49.
| Author | Year | Disease | Classifier | Dataset | Performance metrics |
| Achi et al . 52 | 2019 | lymphoma | CNN | Images | Accuracy 100% |
| Steinbuss et al . 53 | 2021 | lymphoma | CNN | Images | Accuracy 95.56% |
| Li et al . 54 | 2020 | lymphoma | CNN | WSI | Accuracy 100% |
Lymphoma
A class of diseases known as lymphomas is caused by malignant lymphocytes that accumulate in lymph nodes and other lymphoid tissue, resulting in the clinical sign of lymphadenopathy. Based on the histological presence of Reed-Sternberg (RS) cells in Hodgkin lymphoma, lymphomas are primarily classified as either non-Hodgkin lymphoma (NHL) or Hodgkin lymphoma (HL)55. It is difficult to diagnose and subtype NHL; information on these factors is needed in addition to morphological, clinical, serological, and possibly cytogenetic/molecular data54, 53.
Achi et al. (2019) developed a model to employ whole slide imaging (WSI) to distinguish between four forms of lymphomas: diffuse large B-cell lymphoma, small lymphocytic lymphoma, Burkitt lymphoma, and benign lymph node (Table 3). The CNN model employed as a classifier was trained, validated, and tested using 2560 images from 128 cases, 32 of which were of each category. The outcomes showed how effectively the system could detect and classify these classes, with up to 100% accuracy. The study's strength is that it validated the system's performance by separating certain data points and analyzing the model only after it had been trained52.
Furthermore, nodal diffuse large B-cell lymphoma (DLBCL), nodal small lymphocytic lymphoma/chronic lymphocytic leukaemia (SLL/CLL), and tumor-free lymph nodes (LNs) are the subjects of research by Steinbuss et al. (2021). The deep learning model (CNN) achieved an accuracy of 95.56% on the 16,960 test set that was isolated for testing (Table 3). They perform data augmentation following the WSI annotation due to inadequate data. This study shows that it is possible to categorize different forms of lymphoma using AI and differentiate them from other blood malignancies53.
Another use of deep learning, which is CNN, was conducted by Li et al. (2020) (Table 3). They created a highly accurate platform with many CNNs using a small dataset. Pathological images of diffuse large B-cell lymphoma (DLBCL) and non-DLBCL from three hospitals were categorized. Utilizing various data sources is a positive step towards this study's goal of overcoming this challenge. The results indicate that more than 99% accuracy is maintained; however, the performance of AI models in cross-hospital tests decreases after eliminating the technical variability introduced by slide preparation and image collection54.
| Author | Year | disease | Classifier | Dataset | Performance metrics |
| Wei et al . 56 | 2021 | MM | GBDT, DNN, RF and SVM | Tabular data (blood tests) | ROC GBDT-0.9749 RF-0.9693 SVM-0.9052 DNN-0.8696 |
| Chandradevan et al . 57 | 2019 | MM | CNN | BMA smears | ROC Detection-(0.959 ± 0.008) Classification-(0.982 ± 0.03) |
Multiple Myeloma
The neoplastic condition known as Multiple Myeloma (MM) is characterized by an accumulation of plasma cells in the bone marrow, the presence of monoclonal protein in the blood and/or urine, and associated tissue destruction in symptomatic individuals. Therefore, increased clonal plasma cells in the bone marrow, monoclonal protein in serum and/or urine, and related organ or tissue damage are used to diagnose MM55, 58.
Two of the several studies that researchers have conducted to identify and classify MM based on a variety of tabular and image data are used in this study (Table 4). Using tabular data from 4187 regular blood test records, such as serum creatinine, haemoglobin, serum calcium, immunoglobulin (IgG, IgA, IgM), albumin, total protein, and the ratio of albumin to globulin data, Wei et al. (2021) suggested a method to diagnose multiple myeloma (MM). Four methods were used to evaluate the data: GBDT (Gradient Boosting Decision Tree), CNN (Deep Neural Networks), RF, and SVM; the assessment was done using receiver operating characteristic (ROC). The highest ROC value was 0.9749 for GBDT, and 0.9693, 0.9052, and 0.8696 for RF, SVM, and DNN, respectively. These findings enable the use of AI in MM by utilizing information from standard blood tests56.
Furthermore, because haematological diseases may influence the bone marrow, this research counted the cells in the marrow to differentiate multiple myeloma from AML. As a result, Chandradevan et al. (2019) are working on two algorithms: the VGG16 convolutional network for classification and the Faster Region-Based Convolutional Network for detection. High detection (0.959 ± 0.008 precision-recall AUC) and classification (0.982 ± 0.03 ROC AUC) accuracy were shown by the algorithms. To create and evaluate software algorithms, the Wright-stained bone marrow smears (BMA) of seventeen patients were obtained and transformed into whole-slide images, along with three AML and two MM instances. The selection of these illness cases was based on the proportion of malignant cells (20 – 30%) in MM and 30 – 50% in AML.
Numerous studies have used a variety of methods, such as flow cytometry, genetic expression, or medical image recognition, to predict the diagnosis of haematological malignancies. Nevertheless, no research in the literature has ever predicted or detected a blood problem using just the patient's CBC test findings. The latter may be a productive avenue to explore in the future since it is often recognized as the first diagnostic procedure used by haematologists to confirm a leukaemia diagnosis57.
| Author | Year | Disease | Classifier | Dataset | Performance metrics |
| Lotfi et al . 59 | 2015 | IDA | NNET, SVM, KNN With (Maximum Voting theory) | Images of PBS | Accuracy 99%- dacrocyte 97%- elliptocyte 100%- schistocyte |
| Megha et al . 60 | 2016 | IDA | ANN | Images of PBS | Accuracy 81%-Elliptocyte 77%-Dacrocyte 78%-Degmacyte 75%-Schistocyte |
| Betül Çil et al . 2 | 2020 | IDA, Thalassaemia | LR, KNN, SVM | Images of PBS | Accuracy 96.30%-females 94.37%-males 95.95%-both sexes |
| Appiahene et al . 61 | 2023 | IDA | CNN, KNN, NB, DT and SVM | Palm images | Accuracy 99.92%-CNN 99.92%-KNN 99.96%-NB 99.29-DT 96.34%-SVM |
Anaemia
Anaemia is a haematological condition characterized by low RBC and haemoglobin levels. Anaemia is diagnosed through microscopic inspection of a blood smear, which provides detailed information about the size and structure of RBCs. Aside from RBC shape, various tests, such as CBC, contain a wide range of characteristics that can help detect anaemia. AI techniques for detecting anaemia have shown promising results, as detailed in the sections below.
Iron Deficiency Anaemia (IDA)
IDA is the most prevalent haematological disorder caused by a deficiency in iron, which is required for haemoglobin formation and oxygen transport in the body. Reduction in iron concentration causes a decrease in the number of RBCs and alteration in their shape and size. A few studies have reported on the use of ML for IDA diagnosis (Table 5)59. Lotfi et al. (2015) constructed a model to detect poikilocyte cells (dacrocyte, elliptocyte, and schistocyte) in blood smears, which are related to the diagnosis of IDA. The Otsu approach was employed in the segmentation process, while Neural Network (NN), SVM, and K-NN were used as classifiers for the classification process. The proposed algorithm achieved 99%, 97%, and 100% accuracy in detecting dacrocyte, elliptocyte, and schistocyte cells, respectively59. Moreover, Megha et al. (2016) built a model to classify four poikilocyte cells: degmacyte, dacrocyte, schistocyte, and elliptocyte using ANN. The results indicated 81%, 77%, 78%, and 75% accuracy for elliptocyte, dacrocyte, degmacyte, and schistocyte, respectively60.
Due to the similarity in symptoms and testing outcomes between IDA and thalassemia, AI applications have been proposed to distinguish between the two. For example, Betül Çil et al. (2020) proposed a model to differentiate between IDA and thalassemia using CBC parameters from 342 patients. The authors used Logistic Regression (LR), KNN, SVM, Extreme Learning Machine (ELM), and Regularised Extreme Learning Machine (RELM) to detect IDA. The algorithms that produced the most accurate results, categorized by gender, were ELM and RELM (96.30% accuracy for females) and SVM (94.37% accuracy for males). RELM attained an accuracy rate of 95.95% across both genders2.
IDA is typically diagnosed through microscopic examination. However, Appiahene et al. (2023) employed palm images to detect iron deficiency as anaemia causes paleness in many parts of the body. First, the palm images were extracted and augmented before the selection of the region of interest (ROI). Then, the image was processed in CIE L*a*b* color space for feature selection. Using five classifiers for the classification of data (CNN, KNN, NB, DT, and SVM), it was found that CNN performed the best with 99.92% accuracy, whereas SVM achieved the lowest accuracy of 96.34%61.
| Author | Year | disease | Classifier | Dataset | Performance metrics |
| Purwar et al . 19 | 2021 | Thalassaemia | NB, RF, KNN | Images of PBS | 99%-accuracy 100%-specificity 100%-sensitivity |
| Tyas et al . 62 | 2020 | Thalassaemia | MLP | Images of PBS | 98.11%-accuracy |
| Ashari et al . 63 | 2020 | Thalassaemia | RF | Tabular data (CBC parameters) | 100%-accuracy, precision, and recall |
| Author | Year | Disease | Classifier | Dataset | Performance metrics |
| Patgiri et al . 39 | 2021 | SCA | NB, KNN | Images of PBS | Accuracy 98.87%-KNN 98.21%-NB |
| Sen et al . 11 | 2021 | SCA | RF, LR, NB, SVM | Images of PBS | Accuracy 92%-RF 90%-LR 90%-NB 88%-SVM |
| Haan et al . 64 | 2020 | SCA | Images of PBS | 98%-accuracy |
Thalassemia
Thalassemia is a genetic disease that occurs due to a mutation in the haemoglobin (Hb) gene responsible for the synthesis of haemoglobin in RBCs. The Hb gene contains two protein chains, alpha (α) and beta (β). Mutation in either of these genes will lead to a limited supply of oxygen to the organs, making a delayed diagnosis potentially life-threatening65. Recent developments in AI can expedite the verification of the diagnosis (Table 6)19. Purwar et al. (2021) proposed a model to detect thalassemia using clinical reports and images from blood smears. A CNN model was applied to extract features from images and blood analyzers for clinical data. The extracted features were combined and inputted through three classifiers: NB, RF, and KNN. The combined models achieved 99% accuracy, 100% specificity, and 100% sensitivity to classify thalassemic and healthy samples19. However, a perfect score on the parameters can indicate a data overfitting issue, which is discussed in the next section.
Furthermore, Tyas et al. (2020) detected nine abnormal erythrocytes in blood smears of thalassemia patients using multi-layer perceptron (MLP) neural networks. Morphological features, including texture and color features, were extracted from images through the segmentation process in this study. The model achieved 98.11% accuracy in differentiating between thalassemic and healthy samples62. Moreover, tabular data has also been analyzed using AI methods in several studies. For instance, Ashari et al. (2020) used RF to classify thalassemia disease using haematological data. From ten CBC parameters, the model determined haematocrit as the most important variable, and RF analysis showed 100% for all the metrics they used in the study63.
Sickle Cell Anaemia (SCA)
SCA is an inherited haematological disorder caused by an alteration in the haemoglobin gene. Sickle-shaped RBCs, as a result of this disease, have restricted movement in the bloodstream, hence leading to a decrease in oxygen concentration in the body. Various AI methods have been proposed to detect SCA (Table 7)11, 39, 66, 64. For instance, Patgiri et al. (2021) used a hybrid segmentation method called fuzzy C-means and adaptive (local) thresholding for segmentation purposes using ten microscopic blood images to detect SCA. After feature extraction, eight features were used to train classifiers NB and KNN. KNN was found to be slightly more accurate than NB with 98.87% and 98.21% accuracy, respectively39. Furthermore, Sen et al. (2021) classified the cells into three types based on the cells’ morphology: circular (normal cells), elongated (sickle cells), and other cells. The proposed model detected SCA using RF, LR, NB, and SVM and achieved an accuracy of 92%, 90%, 90%, and 88% respectively11.
Additionally, Haan et al. (2020) developed a deep-learning model to diagnose SCA using a blood smear image captured using a smartphone microscope. The structure of the model depends on two NN; the first NN works on improving the quality of the image to be identical to the laboratory microscope image, while the second NN utilizes the enhanced image to differentiate between sickle cells and normal cells. The proposed method achieved an accuracy of 98% when applied to 96 patient samples, including 32 samples taken from SCA patients64.
Challenges in Using AI for the Diagnosis of Haematological Disorders
The use of AI in disease diagnosis and classification is becoming increasingly widespread; however, researchers continue to face several challenges, some of which are discussed in this section.
Data Availability
The availability and accessibility of data are among the main issues facing researchers worldwide because AI requires a considerable amount of data67. In medicine, the lack of testing hampers the rapid growth of AI applications in certain diseases. For instance, AML disease can be diagnosed before a bone marrow biopsy is performed, but while researchers would like to employ AI on bone marrow samples, there is insufficient data on bone marrow images. Furthermore, the ethical concerns and preservation costs associated with sharing patients’ clinical data restrict medical centers and hospitals from contributing their information to public databases68.
Most importantly, the rapid growth of AI applications requires additional steps to validate the initial training datasets. Every supervised model is typically built and optimized using a considerable amount of data known as the 'training set' before conducting tests on a separate ‘testing dataset’. It is often required to validate using another collection of data, known as the ‘validation set’ or the original data set itself after model development6. Consequently, this emphasizes the necessity for enhanced accessibility to the necessary data. However, the issue with inadequate data can now be resolved through data augmentation, for which researchers employ a variety of techniques, such as image preprocessing, rotation, flipping, and sharing, to increase the size of the dataset and prevent memorization—particularly in deep learning algorithms69, 70.
Differences Between Making a Diagnosis and the Quality of Data
AI models require high-quality images and data for processing; however, obtaining a high-quality image could be complicated due to the existence of impurities in the image, imaging using low-resolution microscopes, or incorrect imaging parameters. The variance in quality in the sample microscopic slides could be caused by several reasons, including inaccurate smearing technique, mishandling of the sample, and the use of low-quality reagents such as stains32. One way of overcoming this is by obtaining a significant amount of data from many sources68. However, the variation in images and quality may generate “confusion” in the AI model as the training dataset will not have a coherent pattern. For example, in the case of diagnosing AML, some of the blast cells are indistinguishable, which may result in different annotations by different pathologists. Hence, taking the data from several sources can affect the AI model's accuracy, which can be overcome by re-annotating the data.
Algorithm Challenges
AI methods require a substantially extensive amount of data for the training set, particularly in DL methods. As a result, researchers must resort to techniques augmenting images using specific techniques, which reduces the percentage of accuracy in the result. An additional limitation of AI is that its models are incapable of comprehending situations that require explanations, or "black boxes" as they are referred to in CNN52. For example, pathologists require accurate interpretations of medical diagnoses; however, it should be noted that medically recognized disorders are frequently multidimensional, and there are cases when the reasoning is beyond human comprehension. Thus, techniques for dissecting these "black box" models have been created, which may assist in minimizing the uncertainty56. Lakkaraju et al. (2017) offer a framework called “Black Box Explanations by Transparent Approximations” (BETA), a unique model to explain the behavior of any black box classifier by optimizing the fidelity to the original model and explanation interpretability at the same time. This approach enables the user to explore the black box behavior in different subspaces of their interest, thus eliminating the potential biases of the original model57.
Ethical and Social Implications
Generally, the integration of AI in healthcare has multiple caveats when considering data protection, informed consent, the humane aspect of medical consultations, and the social gaps which could lead to significant bias if AI is trained with non-representative datasets71, 72, 73. Although, in the context of diagnosis of haematological and haemato-oncological disorders, where the primary dataset comprises images of blood smear microscopic slides and numbers from a CBC report, it poses less risk of certain social and ethical implications as compared to using patients’ data in a field like genetic disorders where more sensitive data is involved.
However, regardless of the sensitivity of the information used, regulatory frameworks and bodies must be established to ensure the use of AI with minimal compromise on the ethical and social front. For example, it needs to be ensured that the socioeconomic gaps in our societies do not restrict access to AI so that the trained AI models can account for all variations and not lead to incorrect diagnoses. This is even more significant since blood disorders vary significantly across different ethnic and demographic groups74.
Conclusion
In this article, we reviewed different AI models and their application in the diagnosis of some haematological disorders. Advancements in this technology will not only speed up the process and increase the accuracy of clinical results, but they will also reduce healthcare costs significantly. AI has the potential to bring significant advancement in revolutionizing our healthcare systems. In certain circumstances, however, human intervention and expertise are still necessary to comprehend concepts that lie beyond the capabilities of AI as it can only process and analyze patterns and correlations, but not the context of the information. Researchers may face challenges as AI requires a large amount of data, leading to questions about its quality and integrity; however, this would be subdued when more data is made available. Regulatory frameworks set and enforced by appropriate bodies can be established to minimize the social and ethical implications of AI. As a roadmap for the future, the capabilities of this technology should be explored further in other areas such as personalized medicine, drug discovery, and medical imaging analysis as it has the potential to streamline numerous processes in our healthcare systems.
Abbreviations
AA - Aplastic Anemia, AI - Artificial Intelligence, ALL - Acute Lymphoblastic Leukemia, AML - Acute Myeloid Leukemia, ANN - Artificial Neural Network, BMA - Bone Marrow Aspirate, BM - Bone Marrow, CBC - Complete Blood Count, CLL - Chronic Lymphocytic Leukemia, CML - Chronic Myeloid Leukemia, CNN - Convolutional Neural Network, DLBCL - Diffuse Large B-Cell Lymphoma, DL - Deep Learning, DNN - Deep Neural Network, DT - Decision Tree, FAB - French-American-British (classification system), Hb - Hemoglobin, HL - Hodgkin LymphomaI, DA - Iron Deficiency Anemia, IgG, IgA, IgM - Immunoglobulins G, A, and M, KNN - K-Nearest Neighbor, MDS - Myelodysplastic Syndromes, ML - Machine Learning, MM - Multiple Myeloma, NB - Naïve Bayes, NHL - Non-Hodgkin Lymphoma, PBS - Peripheral Blood Smears, RF - Random Forest, RNN - Recurrent Neural Network, ROC - Receiver Operating Characteristic, SCA - Sickle Cell Anemia
Acknowledgments
None.
Author’s contributions
HA, and AI contributed to the design of the research. HA and ZA extracted the data, analyzed and summarized it. HA, ZA and HNMN drafted the manuscript. AI, MA, HY, NY edited and reviewed. AI, MA, HY supervised the research.All authors read and approved the final manuscript.
Funding
We would like to acknowledge the financial support provided by Universiti Sains Malaysia (USM), Malaysia, under the Short-Term Grant (STG) Scheme (STG Code: 304.CIPPT.6315621).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Amin
M.M.,
Kermani
S.,
Talebi
A.,
Oghli
M.G.,
Recognition of acute lymphoblastic leukemia cells in microscopic images using k-means clustering and support vector machine classifier. Journal of Medical Signals and Sensors.
2015;
5
(1)
:
49-58
.
View Article PubMed Google Scholar -
Çil
B.,
Ayyıldız
H.,
Tuncer
T.,
Discrimination of β-thalassemia and iron deficiency anemia through extreme learning machine and regularized extreme learning machine based decision support system. Medical Hypotheses.
2020;
138
:
109611
.
View Article PubMed Google Scholar -
Yap
M.H.,
Pons
G.,
Martí
J.,
Ganau
S.,
Sentís
M.,
Zwiggelaar
R.,
Moi Hoon Yap
Automated Breast Ultrasound Lesions Detection Using Convolutional Neural Networks. IEEE Journal of Biomedical and Health Informatics.
2018;
22
(4)
:
1218-26
.
View Article PubMed Google Scholar -
van Melle
W.,
MYCIN: a knowledge-based consultation program for infectious disease diagnosis. International Journal of Man-Machine Studies.
1978;
10
(3)
:
313-22
.
View Article Google Scholar -
Gunčar
G.,
Kukar
M.,
Notar
M.,
Brvar
M.,
Černelč
P.,
Notar
M.,
An application of machine learning to haematological diagnosis. Scientific Reports.
2018;
8
(1)
:
411
.
View Article PubMed Google Scholar -
Muhsen
I.N.,
Shyr
D.,
Sung
A.D.,
Hashmi
S.K.,
Machine Learning Applications in the Diagnosis of Benign and Malignant Hematological Diseases. Clinical Hematology International.
2020;
3
(1)
:
13-20
.
View Article PubMed Google Scholar -
Agius
R.,
Brieghel
C.,
Andersen
M.A.,
Pearson
A.T.,
Ledergerber
B.,
Cozzi-Lepri
A.,
Machine learning can identify newly diagnosed patients with CLL at high risk of infection. Nature Communications.
2020;
11
(1)
:
363
.
View Article PubMed Google Scholar -
Walter
W.,
Pohlkamp
C.,
Meggendorfer
M.,
Nadarajah
N.,
Kern
W.,
Haferlach
C.,
Artificial intelligence in hematological diagnostics: game changer or gadget?. Blood Reviews.
2022;
58
:
101019
.
View Article PubMed Google Scholar -
Institute of Electrical and Electronics Engineers, Manav Rachna International Institute of Research and Studies. Proceedings of the International Conference on Machine Learning, Big Data, Cloud and Parallel Computing : trends, prespectives and prospects : COMITCON-2019 : 14th-16th February, 2019 .
.
-
Samuel
A.L.,
Some studies in machine learning using the game of checkers. IBM Journal of research and development.
1959;
3
(3)
:
210-29
.
-
Sen
B.,
Ganesh
A.,
Bhan
A.,
Dixit
S.,
Goyal
A.,
Machine learning based diagnosis and classification of sickle cell anemia in human RBC.. Third International Conference on Intelligent Communication Technologies and Virtual Mobile Networks .
2021;
2021
:
753-8
.
View Article Google Scholar -
Liu
K.,
Hu
J.,
Classification of acute myeloid leukemia M1 and M2 subtypes using machine learning. Computers in Biology and Medicine.
2022;
147
:
105741
.
View Article PubMed Google Scholar -
Ahsan
M.M.,
Luna
S.A.,
Siddique
Z.,
Machine-learning-based disease diagnosis: A comprehensive review. InHealthcare.
2022;
10
(3)
:
541
.
View Article Google Scholar -
Han
J.,
Pei
J.,
Tong
H.,
Data Mining: Concepts and Techniques. 4th ed. Morgan Kaufmann, 2022. 2022
.
-
Shetty
S.H.,
Shetty
S.,
Singh
C.,
Rao
A.,
In Fundamentals and Methods of Machine and Deep Learning, P. Singh (Ed.). https://doi.org/10.1002/9781119821908.ch1. 2022;
1
.
-
Quinlan
J.R.,
Probabilistic decision trees. In: Machine Learning. Elsevier. 1990;
:
140-52
.
-
Goutam
D.,
Sailaja
S.,
Classification of Acute Myelogenous Leukemia in Blood Microscopic Images using Supervised Classifier [Internet]. Available from: www.ijert.org. In2015 IEEE International Conference on Engineering and Technology.
2015;
:
1-5
.
-
Patel
H.,
Prajapati
P.,
Patel
H.H.,
Study and Analysis of Decision Tree Based Classification Algorithms. International Journal of Computer Sciences and Engineering .
2018;
6
(10)
:
74-8
.
View Article Google Scholar -
Purwar
S.,
Tripathi
R.,
Ranjan
R.,
Saxena
R.,
Classification of thalassemia patients using a fusion of deep image and clinical features.. Proceedings of the Confluence.
2021;
:
410-5
.
View Article Google Scholar -
Docking
T.R.,
Parker
J.D.,
Jädersten
M.,
Duns
G.,
Chang
L.,
Jiang
J.,
A clinical transcriptome approach to patient stratification and therapy selection in acute myeloid leukemia. Nature Communications.
2021;
12
(1)
:
2474
.
View Article PubMed Google Scholar -
Gupta
C.,
Saha
A.,
Reddy
N.V.,
Acharya
U.D.,
Cardiac Disease Prediction using Supervised Machine Learning Techniques. nJournal of Physics: Conference Series 2022.
2022;
2161
(1)
:
012013
.
-
Zhang
Q.,
Convolutional Neural Networks.. 3rd International Conference on Electromechanical Control Technology and Transportation.
2018;
:
434-9
.
View Article Google Scholar -
Chauhan
D.,
Walia
R.,
Singh
C.,
Deivakani
M.,
Kumbhkar
M.,
Professor
A.,
Detection of Maize Disease Using Random Forest Classification Algorithm. Turkish Journal of Computer and Mathematics Education.
2021;
12
(9)
:
715-20
.
-
Breiman
L.,
Random Forests. Machine Learning.
2001;
45
:
5-32
.
View Article Google Scholar -
Octaviani
T.L.,
Rustam
Z.,
Random forest for breast cancer prediction. InAIP Conference Proceedings.
2019;
2168
(1)
:
020050
.
View Article Google Scholar -
Arfiani
A.,
Rustam
Z.,
Ovarian cancer data classification using bagging and random forest. InAIP Conference Proceedings.
2019;
2168
(1)
:
020046
.
View Article Google Scholar -
Dasariraju
S.,
Huo
M.,
McCalla
S.,
Detection and classification of immature leukocytes for diagnosis of acute myeloid leukemia using random forest algorithm. Bioengineering (Basel, Switzerland).
2020;
7
(4)
:
1-12
.
View Article PubMed Google Scholar -
Mirmohammadi
P.,
Ameri
M.,
Shalbaf
A.,
Recognition of acute lymphoblastic leukemia and lymphocytes cell subtypes in microscopic images using random forest classifier. Physical and Engineering Sciences in Medicine.
2021;
44
(2)
:
433-41
.
View Article PubMed Google Scholar -
Gaye
B.,
Zhang
D.,
Wulamu
A.,
Improvement of Support Vector Machine Algorithm in Big Data Background. Mathematical Problems in Engineering.
2021;
2021
(1)
:
5594899
.
View Article Google Scholar -
MoradiAmin
M.,
Memari
A.,
Samadzadehaghdam
N.,
Kermani
S.,
Talebi
A.,
Computer aided detection and classification of acute lymphoblastic leukemia cell subtypes based on microscopic image analysis. Microscopy Research and Technique.
2016;
79
(10)
:
908-16
.
View Article PubMed Google Scholar -
Vogado
L.H.,
Veras
R.M.,
Araujo
F.H.,
Silva
R.R.,
Aires
K.R.,
Leukemia diagnosis in blood slides using transfer learning in CNNs and SVM for classification. Engineering Applications of Artificial Intelligence.
2018;
72
:
415-22
.
View Article Google Scholar -
Dese
K.,
Raj
H.,
Ayana
G.,
Yemane
T.,
Adissu
W.,
Krishnamoorthy
J.,
Accurate Machine-Learning-Based classification of Leukemia from Blood Smear Images. Clinical Lymphoma, Myeloma & Leukemia.
2021;
21
(11)
:
e903-14
.
View Article PubMed Google Scholar -
Patrick
E.A.,
Fischer
F.P.,
A generalized k-nearest neighbor rule. Information and Control.
1970;
16
(2)
:
128-52
.
View Article Google Scholar -
Institute of Electrical and Electronics Engineers, Manav Rachna International Institute of Research and Studies. Proceedings of the International Conference on Machine Learning, Big Data, Cloud and Parallel Computing : trends, prespectives and prospects : COMITCON-2019 : 14th-16th February, 2019. . 2019
.
-
Saini
R.,
Bindal
N.,
Bansal
P.,
Classification of heart diseases from ECG signals using wavelet transform and kNN classifier. InInternational conference on computing, communication & automation.
2015;
:
1208-1215
.
View Article Google Scholar -
Mahesh
B.,
Machine Learning Algorithms-A Review. International Journal of Science and Research.
2018;
9
(1)
:
381-6
.
-
Pattekari
S.A.,
Parveen
A.,
Prediction system for heart disease using naive bayes. nternational Journal of Advanced Computer and Mathematical Sciences.
2012;
3
(3)
:
290-4
.
-
Subbalakshmi
M.G.,
Tech
M.,
Ramesh
M.K.,
Chinna
M.M.,
Tech
R.M.,
Decision Support in Heart Disease Prediction System using Naive Bayes. Indian Journal of Computer Science and Engineering.
2011;
2
(2)
:
170-6
.
-
Patgiri
C.,
Ganguly
A.,
Adaptive thresholding technique based classification of red blood cell and sickle cell using Naive Bayes Classifier and K-nearest neighbor classifier. Biomedical Signal Processing and Control.
2021;
68
:
102745
.
View Article Google Scholar -
Vembandasamyp
K.,
Sasipriyap
R.R.,
Deepap
E.,
Heart Diseases Detection Using Naive Bayes Algorithm. International Journal of Innovative Science, Engineering & Technology.
2015;
2
(9)
:
441-4
.
-
H. El Achi,
Khoury
J.D.,
Artificial intelligence and digital microscopy applications in diagnostic hematopathology. Cancers.
2020;
12
(4)
:
797
.
View Article Google Scholar -
Shrestha
A.,
Mahmood
A.,
. 2019 Apr 22;:..
Review of deep learning algorithms and architectures. IEEE access.
2019;
7
:
53040-65
.
View Article Google Scholar -
Yang
F.,
Poostchi
M.,
Yu
H.,
Zhou
Z.,
Silamut
K.,
Yu
J.,
Deep Learning for Smartphone-Based Malaria Parasite Detection in Thick Blood Smears. IEEE Journal of Biomedical and Health Informatics.
2020;
24
(5)
:
1427-38
.
View Article PubMed Google Scholar -
Kang
K.W.,
Kim
H.,
Hur
W.,
Jung
J.H.,
Jeong
S.J.,
Shin
H.,
A Proteomic Approach to Understand the Clinical Significance of Acute Myeloid Leukemia-Derived Extracellular Vesicles Reflecting Essential Characteristics of Leukemia. Molecular & Cellular Proteomics.
2021;
20
:
100017
.
View Article PubMed Google Scholar -
Isa
A.,
Dysregulated Metabolism in Cancer. Biomedical Research and Therapy.
2022;
9
(8)
:
5201-8
.
View Article Google Scholar -
Zini
G.,
Artificial intelligence in hematology. Hematology (Amsterdam, Netherlands).
2005;
10
(5)
:
393-400
.
View Article PubMed Google Scholar -
Obstfeld
A.E.,
Hematology and Machine Learning. The Journal of Applied Laboratory Medicine.
2023;
8
(1)
:
129-44
.
View Article PubMed Google Scholar -
Kimura
K.,
Tabe
Y.,
Ai
T.,
Takehara
I.,
Fukuda
H.,
Takahashi
H.,
A novel automated image analysis system using deep convolutional neural networks can assist to differentiate MDS and AA. Scientific Reports.
2019;
9
(1)
:
13385
.
View Article PubMed Google Scholar -
Wang
M.,
Dong
C.,
Gao
Y.,
Li
J.,
Han
M.,
Wang
L.,
A Deep Learning Model for the Automatic Recognition of Aplastic Anemia, Myelodysplastic Syndromes, and Acute Myeloid Leukemia Based on Bone Marrow Smear. Frontiers in Oncology.
2022;
12
:
844978
.
View Article PubMed Google Scholar -
Salam
R.A.,
Hassan
R.,
Cohen
P.R.,
Adams
N.M.,
Berthold
M.R.,
Ismail
W.,
Image segmentation using a hybrid clustering technique and mean shift for automated detection acute leukaemia blood cells images. Journal of Theoretical & Applied Information Technology.
2015;
76
(1)
:
88-96
.
-
Bennett
J.M.,
Catovsky
D.,
M.T. Daniel,
G. Flandrin,
D.A. Galton,
H.R. Gralnick,
C. Sultan,
Proposals for the classification of the acute leukaemias French‐American‐British (FAB) co‐operative group. British journal of haematology.
1976;
33
(4)
:
451-8
.
View Article Google Scholar -
Steinbuss
G.,
Kriegsmann
M.,
Zgorzelski
C.,
Brobeil
A.,
Goeppert
B.,
Dietrich
S.,
Deep learning for the classification of non-hodgkin lymphoma on histopathological images. Cancers (Basel).
2021;
13
(10)
:
2419
.
View Article PubMed Google Scholar -
Achi
H. El,
Belousova
T.,
Chen
L.,
Wahed
A.,
Wang
I.,
Hu
Z.,
Automated Diagnosis of Lymphoma with Digital Pathology Images Using Deep Learning. Annals of Clinical & Laboratory Science.
2019;
49
(2)
:
153-60
.
-
Li
D.,
Bledsoe
J.R.,
Zeng
Y.,
Liu
W.,
Hu
Y.,
Bi
K.,
A deep learning diagnostic platform for diffuse large B-cell lymphoma with high accuracy across multiple hospitals. Nature Communications.
2020;
11
(1)
:
6004
.
View Article PubMed Google Scholar -
V. Hoffbrand,
G. Collins,
J. Loke,
Hoffbrand's essential haematology. John Wiley & Sons; 2024 .
Google Scholar -
Yan
W.,
Shi
H.,
He
T.,
Chen
J.,
Wang
C.,
Liao
A.,
Employment of Artificial Intelligence Based on Routine Laboratory Results for the Early Diagnosis of Multiple Myeloma. Frontiers in Oncology.
2021;
11
:
608191
.
View Article PubMed Google Scholar -
Alaoui
Y. El,
Elomri
A.,
Qaraqe
M.,
Padmanabhan
R.,
Taha
R.Y.,
Omri
H. El,
A Review of Artificial Intelligence Applications in Hematology Management: Current Practices and Future Prospects. Journal of Medical Internet Research.
2022;
24
(7)
:
e36490
.
-
Allegra
A.,
Tonacci
A.,
Sciaccotta
R.,
Genovese
S.,
Musolino
C.,
Pioggia
G.,
Machine Learning and Deep Learning Applications in Multiple Myeloma Diagnosis, Prognosis, and Treatment Selection. Cancers.
2022;
14
(3)
:
606
.
View Article Google Scholar -
Lotfi
B.N. Mahsa,
The detection of Dacrocyte, Schistocyte and Elliptocyte cells in Iron Deficiency Anemia. International Conference on Pattern Recognition and Image Analysis.
2015;
2015
:
1-5
.
-
Appiahene
P.,
Asare
J.W.,
Donkoh
E.T.,
Dimauro
G.,
Maglietta
R.,
Detection of iron deficiency anemia by medical images: a comparative study of machine learning algorithms. BioData Mining.
2023;
16
(1)
:
2
.
View Article PubMed Google Scholar -
Tyagi
L.M. Megha,
Detection of Poikilocyte Cells in Iron Deficiency Anaemia Using Artificial Neural Network.. International conference on computation of power, energy information and commuincation (ICCPEIC).
2016;
2016
:
108-112
.
-
Tyas
D.A.,
Hartati
S.,
Harjoko
A.,
Ratnaningsih
T.,
Morphological, Texture, and Color Feature Analysis for Erythrocyte Classification in Thalassemia Cases. IEEE Access : Practical Innovations, Open Solutions.
2020;
8
:
69849-60
.
View Article Google Scholar -
Aszhari
F.R.,
Rustam
Z.,
Subroto
F.,
Semendawai
A.S.,
Classification of thalassemia data using random forest algorithm. Journal of Physics: Conference Series 2020 .
2020;
1490
(1)
:
01205
.
View Article Google Scholar -
de Haan
K.,
Ceylan Koydemir
H.,
Rivenson
Y.,
Tseng
D.,
Van Dyne
E.,
Bakic
L.,
Automated screening of sickle cells using a smartphone-based microscope and deep learning. NPJ Digital Medicine.
2020;
3
(1)
:
76
.
View Article PubMed Google Scholar -
Rustam
F.,
Ashraf
I.,
Jabbar
S.,
Tutusaus
K.,
Mazas
C.,
Barrera
A.E.,
Prediction of [Formula: see text]-Thalassemia carriers using complete blood count features. Scientific Reports.
2022;
12
(1)
:
19999
.
View Article PubMed Google Scholar -
Alzubaidi
L.,
Fadhel
M.A.,
Al-shamma
O.,
Zhang
J.,
Duan
Y.,
Deep learning models for classification of red blood cells in microscopy images to aid in sickle cell anemia diagnosis. Electronics (Switzerland).
2020;
9
(3)
:
427
.
View Article Google Scholar -
Shouval
R.,
Fein
J.A.,
Savani
B.,
Mohty
M.,
Nagler
A.,
Machine learning and artificial intelligence in haematology. British journal of haematology.
2021;
192
(2)
:
239-50
.
View Article Google Scholar -
Ting
D.S.,
Peng
L.,
Varadarajan
A.V.,
Keane
P.A.,
Burlina
P.M.,
Chiang
M.F.,
Deep learning in ophthalmology: The technical and clinical considerations. Progress in retinal and eye research.
2019;
72
:
100759
.
View Article Google Scholar -
Ahmed
N.,
Yigit
A.,
Isik
Z.,
Alpkocak
A.,
Identification of leukemia subtypes from microscopic images using convolutional neural network. Diagnostics (Basel).
2019;
9
(3)
:
104
.
View Article PubMed Google Scholar -
Nozaka
H.,
Oda
M.,
Sasaki
A.,
Harako
H.,
Miyazaki
M.,
Kaga
S.,
The effect of data augmentation in deep learning approach for peripheral blood leukocyte recognition. InMEDINFO 2021: One World, One Health–Global Partnership for Digital Innovation.
2022;
2022
:
273-7
.
-
Watson
D.S.,
Krutzinna
J.,
Bruce
I.N.,
Griffiths
C.E.M.,
McInnes
I.B.,
Barnes
M.R.,
Clinical applications of machine learning algorithms: Beyond the black box. Bmj.
2019;
2019
:
364
.
View Article Google Scholar -
Lakkaraju
H.,
Kamar
E.,
Caruana
R.,
Leskovec
J.,
Interpretable & Explorable Approximations of Black Box Models. arXiv preprint arXiv:1707.01154.
2017
.
-
Farhud
D.D.,
Zokaei
S.,
Ethical Issues of Artificial Intelligence in Medicine and Healthcare. Iranian journal of public health.
2021;
50
(11)
:
i-v
.
View Article Google Scholar -
Kirtane
K.,
Lee
S.J.,
Racial and ethnic disparities in hematologic malignancies. Blood, The Journal of the American Society of Hematology.
1699;
130
(15)
:
1699-705
.
View Article Google Scholar
Comments
Article Details
Volume & Issue : Vol 11 No 12 (2024)
Page No.: 6989-7002
Published on: 2024-12-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 1987 times
- PDF downloaded - 492 times
- XML downloaded - 94 times
 Biomedpress
Biomedpress



